Information
Journal Policies
ARC Journal of Immunology and Vaccines
Volume-1 Issue-1, 2016
Abstract
Background: Common variable immunodeficiency (CVID) is the most common primary
immunodeficiency syndrome characterized by hypo-gammaglobulinemia and recurrent bacterial infections. The role of BAFF is to send messages for survival and maturation of B cells through BAFF-R. The aim of the present study was to determine mutations in BAFF-R coding area and transitional B cells frequency among peripheral blood B cells of our registered patients.
Methods: Blood samples were taken from patients with CVID (n=15) and healthy controls (n=15). The percentage of total and transitional B-cells was determined by flow cytometry method, using anti CD19-FITC, anti CD24-PE and anti CD38 PE antibodies. Genomic DNA was extracted and PCR was performed to amplify each of BAFF-R 3 exons and then, the PCR products were sent for sequencing.
Results: The results of the sequence and alignment of the sequences did not show any variation in the patients BAFF-R gene compared with controls. The flow cytometric analysis indicated that the mean percentage of B cells in CVID patients (2.44±0.76, p<0.001) was significantly lower in comparison with control group (10.43±3.46). However, the expression of BAFF-R on B cells as well as the frequency of transitional B cells were not different between the case and control groups.
Conclusion: Our CVID patients showed no mutation in their BAFF-R coding areas. Low B cell count and normal transitional B cells frequency reflects no BAFF-R related defect in our patients.
2.KEYWORDS
3.INTRODUCTION
4.MATERIALS AND METHODS
5.RESULTS
5.1.BAFF-R exons sequence analysis
5.2.The frequency of total and transitional B cells in controls, allergic and non-allergic patients with CVID
6.DISCUSSION
7.ACKNOWLEDGEMENTS
8.REFERENCES
AUTHOR DETAILS
Nahid Eskandari1, Mohammad Alihassanzadeh2, Mazdak Ganjalikhani-Hakemi1*, Roya Sherkat3, Hossein Khanahmad4, Yaghub Yazdani5
1Department of Immunology, Faculty of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran.
2Department of Immunology, Faculty of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran.
3Acquired Immunodeficiency Research Center, Isfahan University of Medical Sciences, Isfahan, Iran.
4Department of Genetics, Faculty of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran.
5Department of Immunology, Faculty of Medicine, Golestan University of Medical Sciences, Golestan, Iran.
*[email protected]
KEYWORDS
CVID, Immunodeficiency, Immunophenotyping, hypogammaglobulinemia
INTRODUCTION
Common variable immune deficiency (CVID) as a heterogeneous group of primary immune deficiencies is characterized by insufficient serum levels of Immunoglobulin's (Igs), reduced response to specific antigens and higher incidence of repeated infections [1, 2]. Autoimmune disorders, lymphoproliferative disorders and gastric complications also have an increased incidence in patients with CVID [3]. After selective deficiency of IgA, CVID is the most common primary immunodeficiency in adult patients with a history of recurrent infection in mucosal surfaces due to reduced serum antibodies [4] with an estimated incidence between 1:10,000 and 1:50,000 [5, 6].The main immunological defect in CVID is failure of B-cells in Ig production, although abnormalities have been described in other components of the immune system [7, 8].
Failure to produce antibodies is attributed to a variety of disorders, including intrinsic problems of B cell, reduced helps from T cells and increased suppressor cell activity. In contrast to most other primary immunodeficiencies, more than 90% of documented CVID patients are lacking a definite molecular genetic diagnosis or other causal explanation for their disease (3, 20). In a minority of patients with CVID, distinct molecular genetic defects have been identified. These genes associated with a CVID phenotype are ICOS (inducible stimulator), TACI (trans membrane activator and calcium-modulating cyclophilin ligand interactor), CD19, BAFF-R, CD81, CD20, CD21 and LRBA (lipopolysaccharide responsive beige-like anchor protein) [9]. The most common cause of CVID and IgA deficiency are associated with a genetic defect in single-pass transmembrane protein named TACI. This molecule is a receptor for the cytokine BAFF that is expressed by follicular dendritic cells (FDCs) and help messages for the stimulation of B-cell activation and survival through TACI and BAFF receptor. A small group of CVID patients has a defect in BAFF receptor [9, 10].
TACI mutations, which are seen in up to 10% of CVID cases, but occur also in 1% of the healthy population, and thus must be regarded as disease modifiers rather than disease-causing gene defects.
BAFF (B cell activating factor), a member of the Tumor Necrosis Factor (TNF) family (also known as TALL-1, THANK, BlyS and zTNF4) and BAFF receptor (BAFF-R) play a fundamental role during the transition from immature T1 to T2 B cells and therefore for the generation of mature B cells in the spleen [11-14]. All mature B cells express BAFF-R on their surface or are able to respond to BAFF [11, 12, 15].
Development of mature B cells is a complex process orchestrated by different gene products. Recent studies have underscored the importance of BAFF and its receptor, BAFF-R, in supporting the survival of the B lymphocytes lineage. BAFF is expressed on the surface of macrophages, monocytes, and dendritic cells, and can also bind two other receptors: BCMA and TACI. These two molecules are also receptors for APRIL, another TNF ligand that does not interact with BAFF-R (3, 4).
BAFF-R is a member of the TNF family which is encoded on human chromosome 22q13.1-13.31, and its gene includes 3 exons. This gene encodes for a type III transmembrane protein with a molecular weight of 19 KDa. This protein is expressed on the surface of B cells; its extracellular domain contains four cysteine residues, which is in contrast to all other known TNF family receptors that contain at least six cysteine residues [16].
Defects in the expression of either BAFF (B cell activating factor) or BAFF-R impairs B cell development beyond the immature, transitional type-1 stage and thus, prevents the formation of follicular and marginal zone B cells, whereas B-1 B cells remain unaffected. The expression of BAFF-R on all mature B cells might suggest a role for BAFF-R signaling also for their in vivo maintenance.
Recent studies have demonstrated that mice defective in BAFF-R gene exhibit an altered profile of the B cell pool, a phenotype observed in BLyS knockout mice as well. These features suggest that mutations in this gene may result in humoral immunodeficiency. To test this hypothesis, we aimed to sequence the BAFF-R gene in 15 patients with common variable immunodeficiency (CVID) along with 15 healthy controls in order to find any probable disease causing mutation. We also tried to compare transitional B cell count in the patients with controls.
MATERIALS AND METHODS
The study population was total 15 patients with CVID registered in Immunology clinic of AL Zahra hospital of Isfahan province, and 15 healthy individuals. From each subject, 5 ml whole blood was taken after completing the questionnaire and the informed consent.
Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque™ (Lymphoprep, Oslo, Norway) density gradient.
The genomic DNA was extracted form samples using Genomic DNA Extraction Kit (Genet bio, South Korea) based on manufacturer's instructions. Extracted DNA was used as template for amplifying all 3 exons of the BAFF-R gene through polymerase chain reaction using specific primers (Table 1). The PCR master mix was prepared in final volume of 25 µl and reaction was done on a thermal cycler (Bio-rad, USA). The reaction was performed in 1 cycle at 94 °C for 5 min and 30 cycles at 94 °C for 30 s, 60 °C (exon 1), 58 °C (exon 2), and 56 °C (exon 3) for 30 s, 72 °C for 30 s and a final extension time at 72 °C for 6 min. The PCR products were analyzed by agarose gel electrophoresis and then, were sent for sequencing (Genfanavaran, Iran). Finally, the sequences were aligned versus reference sequences of each exon using NCBI BLAST.
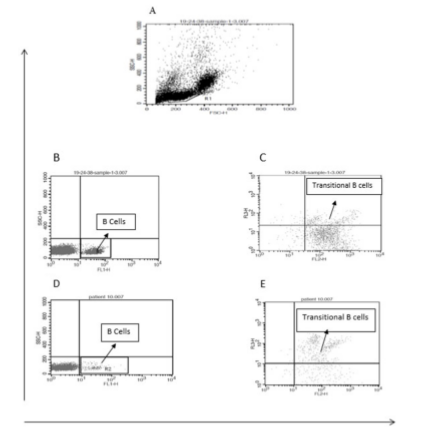
The percentage of transitional B cells were determined using anti-CD19-FITC, anti-CD24-PE and anti-CD38- PerCP (eBiosciences, USA) using the flow cytometery method. Samples were evaluated with FACS Callibour flow cytometer (Becton Dickinson, USA) and the result was analyzed with Cell Quest Pro software.
RESULTS
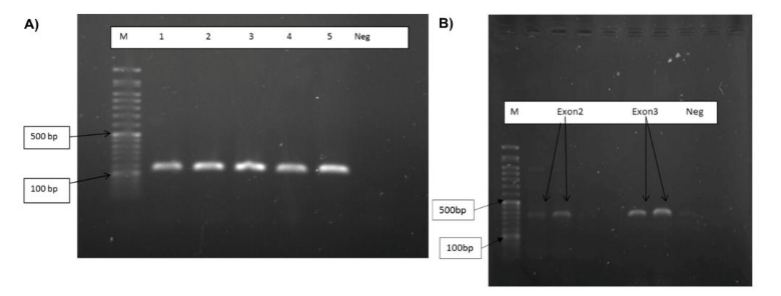
Extracted DNA from patients with CVID and control samples was amplified by specific primers for the 3 exons. Agarose electrophoresis was performed to visualize the PCR products. PCR products in these reactions were 180, 231 and 323bp bands (Figure 1).
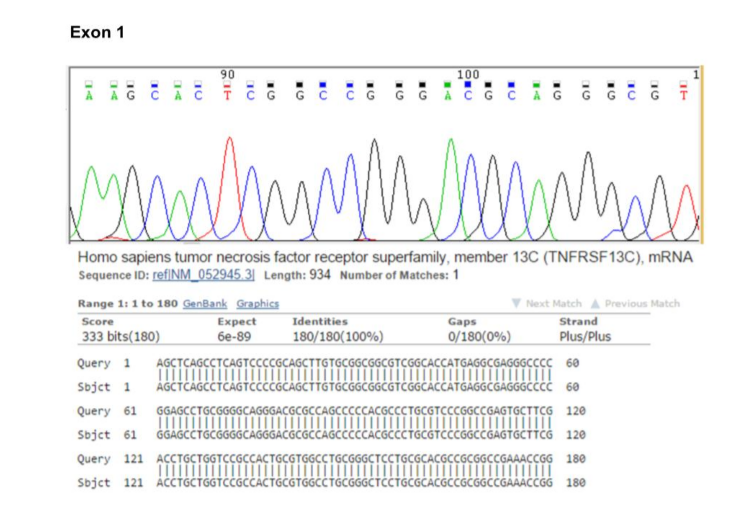
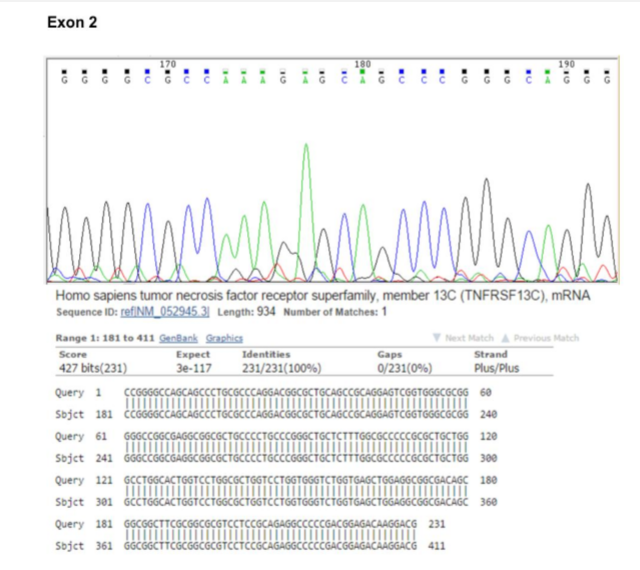
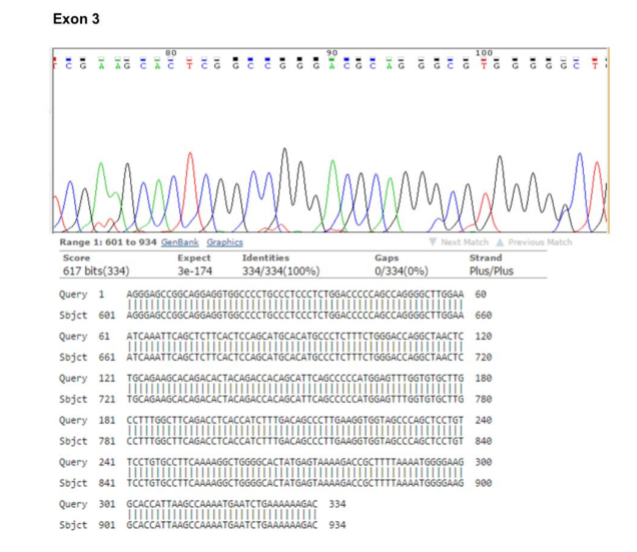
In order to sequence analysis of BAFF-R exons for the possibility of the existence of any mutation, we designed PCR primer pairs that allowed amplification of the 3 exons and their associated flanking splice sites, at least 20 bp upstream and downstream of each exon. After reviewing the results of the sequence and alignment of the sequences, no mutations were observed in BAFF-R exons (Figures 2, 3 & 4).
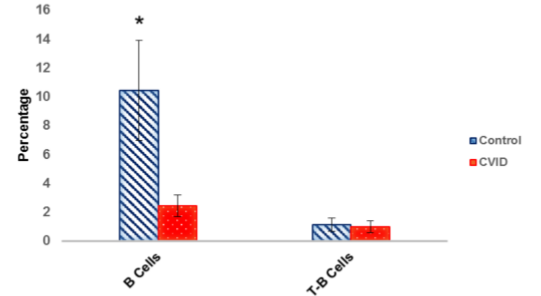
Mean percentage of B cells and transitional B cells were calculated in peripheral blood of 15 patients with CVID and 15 controls using flow cytometry method and is shown in Figure 5. The flow cytometric results indicated that the mean percentage of B cells in control group (10.43±3.46) in comparison with CVID patients (2.44±0.76, p<0.001) was significantly different (Table 2, Figure 6). As shown in Table 2, a difference between the frequency of transitional B cells in control group (1.13±0.45) and CVID patients (0.99±0.4, p=0.8) was seen, but it wasn't statistically significant (Table 2, Figure 6).
DISCUSSION
CVID is a primary immunodeficiency disease characterized by hypo-gammaglobulinemia and recurrent bacterial infections. Although numerous groups have tried to investigate into the nature of the underlying defect, the fundamental cause of this disorder with a few exceptions remains unknown [17]. The phenotypic defect of CVID is a failure in B cell differentiation with impaired secretion of immunoglobulins, which is likely to be caused by different unidentified genetic defects. B cell survival and maturation is a complex process requiring the synergy of numerous factors, which are essential for normal B cell development and function. Recently, B cell activating factor (BAFF, also called BLyS, TALL-1, THANK or zTNF4) and its receptors BAFF-R have been shown to be of great importance for the maturation and maintenance of the peripheral B cells pool by supporting the survival of the B lymphocytes lineage [14, 18, 19]. Recent studies have shown that the three different receptors for BAFF (BCMA, TACI, and BAFF-R) have different and non-redundant functions. BCMA and TACI bind not only BAFF, but also APRIL, whereas BAFF-R remains the only specific ligand for BAFF in B cells [20]. In BAFF knockout mice B cell development is blocked at the transitional type 1 stage (T1). In fact, these mice display a loss of transitional type 2 (T2), follicular (B2), and marginal zone B cells, whereas bone marrow B lymphopoiesis, T1 B cells, B1 cells, and other hematopoietic cell lineages appear unaffected [13]. Conversely, transgenic mice which overexpress BAFF have elevated numbers of peripheral B cells and develop autoimmunity [12].
Also, animal studies, both with knockout and transgenic models, shed light onto the specific functions of each of these receptors. Such studies demonstrated that disruption of BAFF-R, both in the mutant and in the knock-out models, results in an immunological phenotype similar to that observed in CVID, suggesting that BAFF-R might be a good candidate gene for this disorder [11, 13, 21, 22]. Based on different reports and also because of BAFF-R roles in differentiation and maturation of B cells, we hypothesized that there might be any mutation in coding area of BAFF-R gene which could be responsible for at least some cases of CVID. However, our finding showed no mutation in BAFF-R exons on 15 patients with CVID.
In the present study, a reduction of B cells in CVID patients was observed. Similarly, previous studies have reported that in most patients with CVID the frequency of B cells is lower than in normal persons [23]. In addition, we showed no significant difference in the percentage of transitional B cells in patients compared to controls. This finding is correlated with normal BAFF-R gene in our patients as any defect in BAFF-R could cause accumulation of transitional B cells in the patients.
The transitional B cell identification provides a more precise delineation of circulating B cell subsets that will be useful for the evaluation of immunological diseases. Expansion of the transitional B cell compartment has recently been described in HIV patients with advanced diseases [23] and in patients with idiopathic CD4+ T lymphocytopenia [24]. An increased frequency of transitional B cells has also been found in patients with the X linked lymphoproliferative syndrome, a subgroup of common variable immunodeficiency patients and in some patients with systemic lupus erythematosus [25]. Study of lymphocyte subpopulations is an important tool in the diagnosis of immune diseases and also increasingly used to classify patients with CVID into subgroups with different clinical prognosis.
Conclusively, none of our CVID patients showed mutations in their BAFF-R coding areas. Low B cell count and normal transitional B cells frequency reflects no BAFF-R related defect in our patients.
ACKNOWLEDGEMENTS
This work was financially supported by Isfahan University of Medical Sciences (Grant # 392248). We which to thank all individuals, especially patients who took part in this study.
REFERENCES
- Aghamohammadi, A., et al., Clinical and immunological features of 65 Iranian patients with common variable immunodeficiency. Clinical and diagnostic laboratory immunology, 2005. 12(7): p. 825-832.
- Cunningham-Rundles, C. and C. Bodian, Common variable immunodeficiency: clinical and immunological features of 248 patients. Clinical Immunology, 1999. 92(1): p. 34-48.
- Cunningham-Rundles, C., et al., Incidence of cancer in 98 patients with common varied immunodeficiency. Journal of clinical immunology, 1987. 7(4): p. 294-299.
- Olerup, O., et al., Shared HLA class II-associated genetic susceptibility and resistance, related to the HLA-DQB1 gene, in IgA deficiency and common variable immunodeficiency. Proceedings of the National Academy of Sciences, 1992. 89(22): p. 10653-10657.
- Conley, M.E., Diagnostic guidelines-an international consensus document. Clinical Immunology, 1999. 93(3): p. 189.
- Bonilla, F.A., et al., Practice parameter for the diagnosis and management of primary immunodeficiency. Annals of allergy, asthma & immunology, 2005. 94(5): p. S1-S63.
- Oraei, M., et al., 2 Naïve CD4+ T cells and Recent Thymic Emigrants in Common Variable Immunodeficiency. Journal of Investigational Allergology and Clinical Immunology, 2012. 22(3): p. 160.
- Rezaei, N., et al., Cytokine production by activated T cells in common variable immunodeficiency. J. Investig. Allergol. Clin. Immunol, 2010. 20(3): p. 244-251.
- Bacchelli, C., et al., Translational Mini-Review Series on Immunodeficiency: Molecular defects in common variable immunodeficiency. Clinical & Experimental Immunology, 2007. 149(3): p. 401-409.
- Ameratunga, R., et al., New diagnostic criteria for common variable immune deficiency (CVID), which may assist with decisions to treat with intravenous or subcutaneous immunoglobulin. Clinical & Experimental Immunology, 2013. 174(2): p. 203-211.
- Gross, J.A., et al., TACI and BCMA are receptors for a TNF homologue implicated in B-cell autoimmune disease. Nature, 2000. 404(6781): p. 995-999.
- Mackay, F., et al., Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. The Journal of experimental medicine, 1999. 190(11): p. 1697-1710.
- Schiemann, B., et al., An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science, 2001. 293(5537): p. 2111-2114.
- Shulga-Morskaya, S., et al., B cell-activating factor belonging to the TNF family acts through separate receptors to support B cell survival and T cell-independent antibody formation. The Journal of Immunology, 2004. 173(4): p. 2331-2341.
- Batten, M., et al., BAFF mediates survival of peripheral immature B lymphocytes. The Journal of experimental medicine, 2000. 192(10): p. 1453-1466.
- Losi, C.G., et al., Mutational analysis of human BAFF receptor TNFRSF13C (BAFF-R) in patients with common variable immunodeficiency. Journal of clinical immunology, 2005. 25(5): p. 496-502.
- Grimbacher, B., et al., Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nature immunology, 2003. 4(3): p. 261-268.
- Gorelik, L., et al., Cutting edge: BAFF regulates CD21/35 and CD23 expression independent of its B cell survival function. The Journal of Immunology, 2004. 172(2): p. 762-766.
- TNF Family Member B Cell-Activating Factor (BAFF) Receptor-Dependent and-Independent Roles for BAFF in B Cell Physiology.
- Ng, L.G., et al., B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. The Journal of Immunology, 2004. 173(2): p. 807-817.
- Bodmer, J.-L., P. Schneider, and J. Tschopp, The molecular architecture of the TNF superfamily. Trends in biochemical sciences, 2002. 27(1): p. 19-26.
- Thompson, J.S., et al., BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science, 2001. 293(5537): p. 2108-2111.
- Malaspina, A., et al., Appearance of immature/transitional B cells in HIV-infected individuals with advanced disease: correlation with increased IL-7. Proceedings of the National Academy of Sciences of the United States of America, 2006. 103(7): p. 2262-2267.
- Malaspina, A., et al., Idiopathic CD4+ T lymphocytopenia is associated with increases in immature/transitional B cells and serum levels of IL-7. Blood, 2007. 109(5): p. 2086-2088.
- Sims, G.P., et al., Identification and characterization of circulating human transitional B cells. Blood, 2005. 105(11): p. 4390-4398.