Information
Journal Policies
Aetiologically Relevant Typing of E. Coli Isolates from Diseased Pigs in Switzerland during 2014 and 2015
M. Schneeberger, O. Dietz, C. Kirchgässner, S. Schmitt
Copyright : © 2017 Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
In this study, 210 intestinal Escherichia (E.) coli isolates from enteropathogenic diseased pigs were screened for their virulence factors (F4, F5, F6, F18, STa, STb, LT, STx2e and intimin) by PCR and serogrouped by slide-agglutination (O8:K87, O45, O138:K81, O139:K82, O141:K85, O147:K89, O157 and O149:K91). In total, 24.8% enterotoxigenic E. coli, 7.1% enteropathogenic E. coli and 5.2% oedema disease E. coli could be isolated. For enterotoxigenic E. coli, the most frequent virulence pattern was F4/STb/LT with the O-serogroup O149:K91 or O8:K87, followed by F18/STa/STb with the O-serogroup O141:K85. F18/STx2e was the common virulence pattern in oedema disease E. coli isolates and they were associated with the O-serogroups O139:K82 and O141:K85.
Escherichia coli, ETEC, EDEC, EPEC, pig, Switzerland.
1.Introduction
Escherichia (E.) coli is a globally significant pathogen responsible for diarrhoea in neonatal and postweaning piglets as well as for oedema disease. Affected animals show a decreased growth rate and an increased morbidity and mortality rate, resulting in a negative economic effect in swine husbandry [1].
The three significant porcine E. coli pathotypes can be distinguished on the basis of their fimbrial adhesions/proteins and toxins. Enterotoxigenic E. coli (ETEC) are responsible for diarrhoea in neonatal piglets to an age of four days and in postweaning piglets. ETEC colonise the small intestine, as they adhere to the epithelia cells with fimbrial antigens (F4, F5, F6, F18 and F41) and/or an adhesin involved in diffuse adherence (AIDA) [2]. Current studies show that F4 and F18 are the most frequent fimbriae in ETEC strains [3-5]. The heat-stable (STa, STb) and heat-labile (LT) enterotoxins induce diarrhoea while they influence the absorption and secretion of water and electrolytes [6].
Attaching and effacing lesions (A/E) in the small intestine mucosa are linked to enteropathogenic E. coli (EPEC) and affect post weaningpiglets. Their attribute is intimin, a protein which is responsible for the intimate adherence to the host cell. This leads to a reduction in the brush border, which results in malabsorption and finally diarrhoea [6, 7].
In contrast, oedema disease is caused by oedema disease E. coli (EDEC) and usually occurs in postweaning piglets one to two weeks after weaning. The determining factor to classify an EDEC strain is the production of shigatoxin 2e (Stx2e), often associated with the occurrence of the fimbrial adhesin F18 [2, 4, 5]. Unlike enterotoxins, Stx2e can affect endothelial cells, leading to strong vascular damage [2].
Only a limited number of recognized O-serogroups are associated with disease in swine. Both EDEC and ETEC often carry O138, O139, O141, O147 and O157. Additionally, ETEC also frequently carry the O-antigens O8 and O149, whereas EPEC strains often occur in combination with the antigen O45 [2].
Within the intestinal E. coli genepool it is challenging to identify the etiologically relevant E. coli type and to achieve a clear classification into the three swine-relevant pathotypes. This is due to the fact that apathogenic E. coli strains can carry the same O-antigens as the pathogenic strains [8]; haemolytic activity linked with intestinal pathogenicity is discussed controversially in literature [4, 9, 10]; and E. coli carrying virulence factor genes have also been isolated from healthy pigs [8].
To our knowledge, the last study focusing on Swiss E. coli isolates, from diseased piglets, was by Sarrazin et al. [3]. In that study, serogroup O149:K91 with the fimbriae F4 and the enterotoxins LT and STb was dominant. Therefore, the aim of this study was to screen the current situation of pathogenic E. coli in Switzerland and to monitor if there is a shift to another dominant serogroup/pathotype. Additionally, we aimed to develop a rapid, economical and simple diagnostic protocol to reliably detect the pathogenic E. coli isolates out of faecal samples using a multiplex PCR.
2. Materials and Methods
In 2014 and 2015, during a study from the Swiss Federal Food Safety and Veterinary Office, contents from the small intestines were collected during necropsy of swine with diarrhoea.
The samples were cultured on Columbia blood agar containing sheep blood and Gassner agar (both Oxoid, Hampshire, United Kingdom) and incubated overnight at 37 +/–1°C. Afterwards, all colonies suspicious for E. coli were selected. To confirm species identification they were plated on chromogenic agar (Brilliance UTI, Oxoid, Hampshire, United Kingdom). The haemolytic activity was detected on the Columbia blood agar by a zone of complete erythrocyte lysis surrounding the colonies. The E. coli isolates were stored in a cryopreservative solution (pro-lab diagnostics, Bromborough, United Kingdom) at –80°C for further research.
Serogrouping was performed by slide-agglutination with bacteria from overnight pure cultures. Agglutination was primarily performed with the polyspecific testserum ‘Anti-Coli P’ (sifin diagnostics GmbH, Berlin, Germany). This polyspecific testserum contains antibodies against the fimbrial antigen F4 (K88) and a panel of selected O-antibodies (O8:K87, O138:K81, O139:K82, O141:K85, O147:K89 and O149:K91). Additionally, the monospecific testsera O45, O157 (both SSI, Copenhagen, Denmark) and F5 (K99) (sifin diagnostis GmbH, Berlin, Germany) were also used. If an E. coli isolate reacted positively with ‘Anti-Coli P’, the isolate was subsequently tested with the monospecific testsera (sifin diagnostics GmbH, Berlin, Germany) F4, O8:K87, O138:K81, O139:K82, O141:K85, O147:K89 and O149:K91. To check autoagglutination, 0.15 M saline was used as a negative control reagent.
E. coli DNA was isolated from a loopful of bacteria from a fresh Columbia blood agar overnight culture using InstaGene™ Matrix (Bio Rad, Hertfordshire, United Kingdom) and diluted to a working concentration of 10 ng/µl with ddH2O. The DNA was then investigated for presence of the virulence and adhesion genes estA (STa), estB (STb), elt (LT), faeG (F4), fanA (F5), fasA (F6), fedA (F18), eae (intimin) and stx2e (Stx2e) by four separate multiplex PCRs using the primers and controls described in Table 1.
All four multiplex PCRs were performed with the Qiagen HotStarTaq Master Mix (Quiagen, Hilden, Germany) in total reaction volumes of 10 µl: 5 µl HotStarTaq Master Mix, 2 µl ddH2O, 1 µl primer mix (10 µM each), 2 µl DNA sample (10 ng/µl). PCR was performed with following cycling conditions: 1 cycle of 15 min at 95°C; 40 cycles consisting of 30 sec at 94°C, 45 sec at 55°C and 40 sec at 72°C; followed by final elongation for 10 min at 72°C. Resulting PCR products were analysed by automated native capillary electrophoresis (QIAxcel, Qiagen, Hilden, Germany). Functionality of the four multiplex PCRs was validated by analysing 20 E. coli DNA samples previously investigated by singleplex PCR, which was used in the routine diagnostic until now.
3. Results
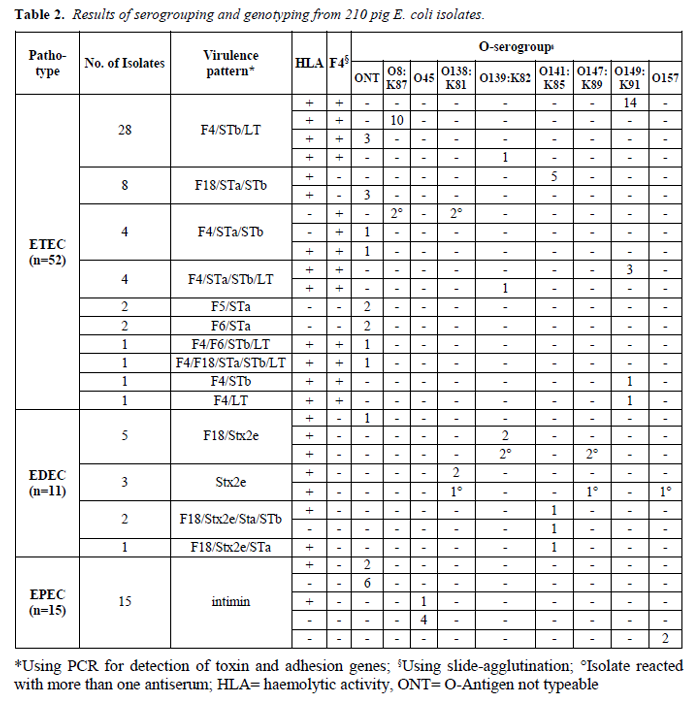
In total, 210 E. coli isolates were screened for their virulence pattern and grouped into the swine-relevant pathotypes ETEC, EPEC and EDEC. Additionally, the serogroup was determined. 78 isolates (37.1%) could be classified in one of the above-named pathotypes (Table 2).
The most common virulence pattern was F4/STb/LT (n=28; 53.8%). All of these isolates showed haemolysis and the dominating serogroup was O149:K91 (n=14), followed by O8:K87 (n=10) and O139:K82 (n=1). Eight isolates harboured the pattern F18/STa/STb and were also haemolytic. Out of this group, five isolates belonged to the serogroup O141:K85.
The virulence pattern F4/STa/STb was detected in four isolates. Three of them were non-haemolytic, whereas one was haemolytic, and two isolates were positive for both O-antigens O8:K87 and O138:K81. Four haemolytic isolates had the pattern F4/STa/STb/LT with the serogroups O149:K91 (n=3) and O139:K82 (n=1). Less frequently detected virulence patterns are summarised in Table 2.
All antigen F4 agglutination-positive E. coli isolates could be confirmed by PCR.
The gene for Stx2e was detected in eleven isolates, of which ten (91%) showed haemolytic activity. The most common virulence pattern was F18/Stx2e (n=5). Two of them belonged to the serogroup O139:K82, whereas two isolates were positive for both O-antigens O139:K82 and O147:K89. In three isolates, only the gene for Stx2e could be detected. Two of them belonged to the serogroup O138:K81, and one isolate reacted with three different antisera (O138:K81, O147:K89 and O157). Another three isolates belonged to the serogroup O141:K85, whose virulence patterns were F18/Stx2e/STa/Stb (n=2) and F18/Stx2e/STa (n=1).
In 15 isolates, the gene coding for the protein intimin could be detected. The dominating serogroup was O45 (n=5), followed by O157 (n=2). For all other isolates, the serogroup could not be determined using our O-antisera test panel. The majority of these isolates were non-haemolytic (n=12; 80%).
Using our investigating panel, 132 isolates (62.9%) were not classifiable. The predominant part (n=108; 81.8%) were non-haemolytic. This heterogeneous group can be classified in four main clusters: i) Serogrouping and ‘virulence genotyping’ negative (n=67; 50.8%), ii) Serogrouping positive but ‘virulence genotyping’ negative (n=42; 31.8%), iii) Isolates carrying enterotoxin genes and partly O-antigen-positive (n=21; 15.9%), iv) Isolates harbouring adhesion genes and partly O-antigen-positive (n=2; 1.5%). In clusters ii, iii and iv, all investigated serogroups occurred, and multiple O-antigen agglutination with monovalent sera often arose.
4. Discussion
In total, 210 E. coli isolates from small intestine probes of piglets with diarrhoea were analysed whether they could be grouped into one of the swine-relevant pathotypes (ETEC, EDEC, and EPEC). In total, 78 out of 210 E. coli isolates (37.1%) could be classified in one of the above-named pathotypes.
ETEC was the most represented pathotype (n=52 isolates; 66.7%) with the dominating virulence pattern F4/STb/LT and the O-serogroup O149:K91. This is in accordance with the Swiss situation about 15 years ago [3] and also with other studies from Europe and the US [4, 5, 11]. The fimbrial gene for F4 was detected uniquely in ETEC isolates and was the predominant adhesion factor (77%). These findings agree with other studies from different countries [4, 5], where the fimbrial gene for F4 was one of the most frequent genes found in piglets suffering from diarrhoea. In contrast, a study from Mexico and China provided other results, indicating that the presence of virulence genes in E. coli varies with the geographical area [12, 13]. The serogroup O149:K91 was clearly correlated with the F4 fimbrial gene, and contrarily O141:K85 was correlated with the F18 fimbrial gene. These findings substantiate other studies [4, 9]. In the majority of these F4 and F18 fimbrial gene-positive isolates, more than one enterotoxin gene could be detected simultaneously, which is in agreement with the study of Zhang et al.[5]. 86% of these F4 and F18 fimbrial gene-positive isolates showed haemolytic activity. In literature, haemolytic activity linked with intestinal pathogenicity is discussed controversially [4, 9, 10]. However, in this study isolates with a full pattern of virulence genes (adhesion and toxin genes) showed haemolytic activity on blood agar more often than other isolates. Therefore, we assume that haemolytic activity is not a determined virulence factor itself, but it could give a hint for pathogenic E. coli isolates. 46 out of 52 ETEC isolates harboured the STb gene in combination with other enterotoxins. These findings substantiate other studies, where STb was the most frequently detected enterotoxin and often occurred in ETEC isolates in combination with other enterotoxins [3,4], except the study of Toledo et al. [12], where the gene for STa was the most common enterotoxin.
In 73% of the EDEC isolates, genes coding for Stx2e and F18 were detected simultaneously, and 91% of the Stx2e-positive isolates showed haemolytic activity. These findings agree with the study of Zhang et al. [5]. The gene for Stx2e occurred in variable combinations with other genes for enterotoxins, which is in concordance with other studies [4, 5].
The gene coding for intimin was detected in 15 isolates, of which the majority was non-haemolytic and the dominating serogroup was O:45. This is in agreement with other studies [14, 15]. Malik et al. [15] showed that the incidence of intimin-positive strains in diarrhoeal and non-diarrhoeal pigs did not differ significantly. Therefore, the pathogenic significance of intimin-positive isolates in pigs remains questionable and only a combination with histopathology can provide assurance. However, the typical attaching and effecting lesions (A/E) in the small-intestine mucosa are often sparse and have to be searched carefully [6, 7].
Using our investigating panel, 132 (62.9%) of the 210 investigated E. coli isolates were not classifiable. Our study focused primarily on the most common serogroups and virulence/adhesion genes associated with gastroenteric diseases in pigs, and data for other virulence or adhesion factors (e.g. AIDA, PAA, EAST1, F41) [1] are missing. Nevertheless, about half of the non-classifiable isolates (50.8%; n=67) were lacking virulence and adhesion genes and reacted with none of the eight tested antisera. Against the background that multiple virulence factors are necessary for pathogenic E. coli isolates [8], we assume these isolates to be non-pathogenic or a component of the normal intestinal gut flora. Interestingly, in this study we found E. coli isolates, which reacted positively with antisera associated with pathogenic E. coli strains by slide-agglutination, but no virulence factors could be detected. These results suggest that serogrouping itself is not discriminating enough to decide if the isolated E. coli strain is the pathogenic agent. Simultaneous agglutination with more than one testserum occurred in this study. Sting and Stermann [9] excluded isolates with more than one positive reaction from their study, and Garabal et al. [16] and Frydendahl et al. [4] performed an O-antigen titration to identify the dominant serogroup. In the presented study, the isolates with multiple agglutination were included, and interestingly the main part occurred in isolates that lacked any virulence or adhesion genes, leading to the conclusion that these isolates were probably commensals.
5.Conclusion
In view of the prudent use of antimicrobial agents, it is crucial to detect and differentiate pathogenic E. coli isolates from the normal gut flora. Therefore, good diagnostic tools are essential. Slide-agglutination with the F4 antisera and the haemolytic activity can give a clue for a pathogenic ETEC or EDEC isolate, but definitive confirmation should be done using virulence genotyping. Several studies showed that pathogenic E. coli isolates could also be detected in clinically healthy pigs [10, 17] and hence for the final diagnosis, clinical and if available macroscopy/ histopathology findings should also be included.
Acknowledgements
The authors thank the Swiss Federal Food Safety and Veterinary Office for the financial support. They are grateful to Prof. Dr. R. Bauerfeind (Justus-Liebig-Universität, Giessen, Germany) for providing the E. coli strains for positive controls, Natasha Carroli for her excellent technical assistance and the Institute of Veterinary Pathology (Vetsuisse Faculty, University of Zurich, Switzerland) for collecting faecal samples during necropsy.
References
- Fairbrother J.M., Nadeau É., Gyles C.L. (2005). Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev., 6(1): 17–39.
- Fairbrother J.M., Gyles C.L.: Escherichia coli infections. In: Diseases of swine, 9th edition, Ed. B.E. Straw, J.J. Zimmerman, S. D’Allaire, D.J. Taylor, Blackwell Publishing, Ames, 2006, 649–662.
- Sarrazin E., Fritsche C., Bertschinger H.U. (2000). Hauptvirulenzfaktoren bei Escherichia coli-Isolaten von über zwei Wochen alten Schweinen mit Ödemkrankheit und/oder Colidiarrhöe. Schweiz. Arch. Tierheilk., 142: 625–630.
- Frydendahl K. (2002). Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol., 85: 169–182.
- Zhang W., Zhao M., Ruesch L., Omot A., Francis D. (2007). Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet. Microbiol., 123: 145–152.
- Nataro J.P., Kaper J.B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev., 11(1): 142–201.
- DebRoy C., Maddox C.W. (2001). Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev., 1(2): 129–140.
- Schierack P., Steinrück H., Kleta S., Vahjen W. (2006). Virulence Factor Gene Profiles of Escherichia coli Isolates from Clinically Healthy Pigs. Applied Enviro. Microbiolog., 72(10): 6680–6686.
- Sting R., Stermann M. (2008). Duplex real-time PCR assays for rapid detection of virulence genes in E. coli isolated from post-weaning pigs and calves with diarrhoea, Dtsch. Tierärzt. Wschr., 115(6): 231–237.
- Schierack P., Weinreich J., Ewers C., Tachu B., Nicholson B., Barth S. (2011). Hemolytic Porcine Intestinal Escherichia coli without Virulence-Associated Genes Typical of Intestinal Pathogenic E. coli. Appl. Environ. Microbiol., 77(23): 8451–8455.
- Vu-Khac H., Holoda E., Pilipcinec E., Blanco M., Blanco J.E., Dahbi G., Mora A., López C., González E.A., Blanco J. (2007). Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhoea in Slovakia. Vet J., 174: 176–87.
- Toledo A., Gómez D., Cruz C., Carreón R., López J., Giono S., Castro A.M. (2012). Prevalence of virulence genes in Escherichia coli strains isolated from piglets in the suckling and weaning period in Mexico. J. Med. Microbiol., 61: 148–156.
- Liu W., Yuan C., Meng X., Du Y., Gao R., Tang J.T., Shi D. (2014). Frequency of virulence factors in Escherichia coli isolated from suckling pigs with diarrhoea in China. Vet. J., 199: 286– 289.
- An H., Fairbrother J.M., Désautels C., Mabrouk T., Dugourd D., Dezfulian H., Harel J. (2000). Presence of the LEE (locus of enterocytes effacement) in pig attaching and effacing Escherichia coli and characterization of eae, espA, espB and espD genes of PEPEC (pig EPEC) strains 1390. Micro.Pathog., 28: 291-300
- Malik A., Tóth I., Beutin L., Schmidt H., Taminiau B., Dow M.A., Morabito S., Oswald E., Mainil J., Nagy B. (2006). Serotypes and intimin types of intestinal and faecal strains of eae+ Escherichia coli from weaned pigs. Vet. Microb. 114: 82–93.
- Garabal J.I., Gonzalez E.A., Vazquez F., Blanco J., Blanco M., Blanco J.E. (1996). Serogroups of Escherichia coli isolated from piglets in Spain. Vet. Microbiol., 48: 113–123.
- Moredo F.A., Piñeyro P.E., Márquez G.C., Sanz M., Colello R., Etcheverría A., Padola N.L., Quiroga M.A., Perfumo C.J., Galli L., Leotta G.A. (2015). Enterotoxigenic Escherichia coli Subclinical Infection in Pigs: Bacteriological and Genotypic Characterization and Antimicrobial Resistance Profiles. Food borne Pathog. Dis., 12(8): 704–7011.
- Bosworth B.T., Casey T.A. (1997). Identification of toxin and pilus genes in porcine Escherichia coli using polymerase chain reaction (PCR) with multiple primer pairs. 97th General Meeting, Am. Soc. Microbiol., Abstract B–509, page 116.
- Franck S.M., Bosworth B.T., Moon H.W. (1998). Multiplex PCR for Enterotoxigenic, Attaching and Effacing, and Shiga Toxin-Producing Escherichia coli Strains from Calves. J. Clin. Microbiol., 36(6): 1795–1797.