Information
Journal Policies
Bioinformatics Tools Stuty on the Sequence and Structure Characteristics of Urinary Prokallikrein
Li Gun*, Yang Ru, Tian Han, Lu Jingqi
Copyright :© 2017 Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Urinary prokallikrein in human participates in the regulation of water and electrolyte balance. In this paper, several characteristics of urinary prokallikrein are studied via some bioinformatics tools. First, the chemical and physical properties such as the molecular weight, the isoelectric point and the hydrophobicity, etc, of urinary prokallikrein are predicted via ProtParam tool. The results show that the molecular formula is C1168H1764N310O363S14, the instability index is 37.98, the theoretical pI is 4.57, et al. the secondary structure of urinary prokallikrein is predicted via the SOPMA tool, the result shows that there are 57(23.95%) Alpha helix, 60(25.21%) Beta sheet, 31(13.03%) Turn and 90(37.82%) Coil. Finally, phosphorylation sites and the protein-protein interaction of urinary prokallikrein are respectively studied by the netphos tool and the STRING system.
Urinary Prokallikrein; Bioinformatics Analysis; Protein-protein Interaction; Sequence ,Urology
1. Introduction
Takahashi, et al. studied the sequence characteristics of the urinary prokallikrein in 1988, they tended to think that the human urinary prokallikrein and kallikrein are of tissue type and they are excreted in urine without any modification [1]. Irie, et al. studied the reactivities of human urinary prokallikrein toward rabbit antibodies [2]. Kamada, et al. explored the tissue prokallikrein activation by some serine proteases in rat [3]. Binding of tissue prokallikrein to isolated human neutrophils was studied by some people twenty years before [4]. Other questions such as the correlation of two different assays for urinary kallikrein [5],the activation of urinary kallikrein in patients with interstitial cystitis [6], urinary kallikrein and salt sensitivity in essential hypertensive males [7], and so on [8-12] were studied by many people too, some works pointed out that there may be some relations between kidney cortex and urinary kallikrein [13,14]. In this paper, we explore the sequence characteristics of urinary kallikrein via some bioinformatics tools.
2. Materials And Methods
In order to study the urinary prokallikrein from the bioinformatics perspective, the sequences data is downloaded from National Center for Biotechnology Information (NCBI) (accession number: 1409287A). Chemical and physical properties are predicted by ProtParam tool (http://web.expasy.org/protparam/) [15]. The secondary structure of urinary prokallikrein is predicted via the SOPMA tool[16]. Then, the phosphorylation sites and the protein-protein interaction of urinary prokallikrein are respectively studied by the netphos tool and the STRING system [17]. The hydrophobicity of urinary prokallikrein is analyzed by the protscale system [18].
3. Results And Discussion
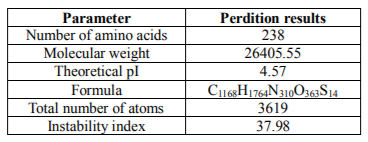
Physical and chemical properties of urinary prokallikrein are studied by protparam system (Table 1). Physical and chemical properties for urinary prokallikrein prediction results show that there are 238 amino acids with the molecular weight is 26405.55. Total number of atoms is 3619, and instability index is 37.98.
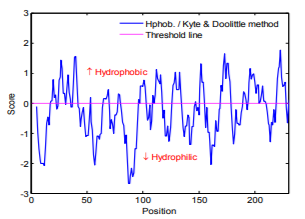
Hydrophobicity is determined by the amino acids sequences. The Hydrophobicity map of urinary prokallikrein sequences is calculated by the Hphoh. / Kyte & Doolittle scale in ExPASy's ProtScale program and the result is shown in Fig.1. In the Fig.1, while vertical value above 0 denotes the hydrophobic area. And it is easily can be seen that there are more hydrophilic residues than the hydrophobic residues in urinary prokallikrein.
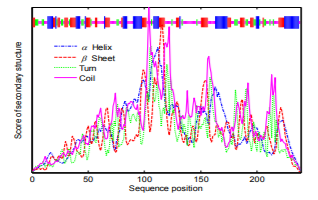
The most common secondary structure is the α-helix, β-fold, turn and random coil. The secondary structure of urinary prokallikrein is predicted via SOPMA system, and the prediction result is shown in Fig.2. The result shows that there are 57(23.95%) alpha helix, 60(25.21%) beta sheet, 31(13.03%) turn and 90(37.82%) coil in the urinary prokallikrein sequence.
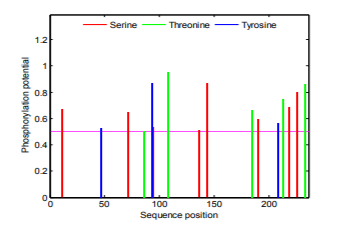
The main sites for protein phosphorylation are serine, threonine and tyrosine. These sites for phosphorylation in urinary prokallikrein are studied via the NetPhos system, and the result is shown in the Fig.3. There are 8 serine phosphorylation sites (potential value higher than 0.5), 5 threonine phosphorylation sites (potential value higher than 0.5), and 3 tyrosine are phosphorylation sites (potential value higher than 0.5) in the urinary prokallikrein.
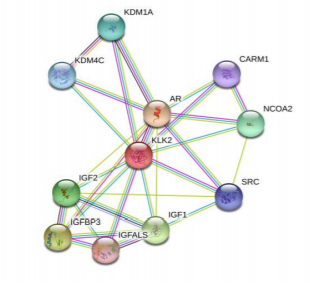
Protein-protein interaction characteristics of urinary prokallikrein is predicted via String online system, and the results is shown in the Fig.4. The results show that there are 11 nodes in the protein-protein interaction network, and there are 29 interaction edges with the average node degree 5.27, PPI enrichment p-value: 0.000146.
The structural characteristics of urinary prokallikrein have not attracted many scientists, so, there is very few related works. In our study, some important characteristics of urinary prokallikrein are predicted via some famous prediction tools, and all of these tools are all widely used in studying the structure of proteins.
4. Conclusion
Although protein techniques can be used for obtaining protein expression data, the bioinformatics tools may be used to verify this data. It is important to investigate information contained in urinary prokallikrein. In this paper,sequence characteristics, such as basic physical and chemical properties, phosphorylation sites, protein-protein interaction network of urinary prokallikrein are predicted via some bioinformatics tools.
Acknowledgments
This research work was supported by president fund of Xi’an Technological University (No.XA GDXJJ14011).
References
- Saori Takahashi, Akiko Irie, and Yoshihiro Miyake. Primary Structure of Human Urinary Prokallikrein. Journal of Biochemistry, 1988, Volume 104, Issue 1, Pages 22-29
- Akiko Irie, Saori Takahashi, Yoshiaki Katayama, Yukio Shibata, Yoshihiro Miyake. Reactivities of human urinary prokallikrein and kallikrein toward rabbit antibodies with special reference to enzyme immunoassay. Clinica Chimica Acta, 1988, Volume 173, Issue 3, Pages 289-297
- Masafumi Kamada, Naoto Furuhata, Takamasa Yamaguchi, Masahiko Ikekita, et al. Observation of tissue prokallikrein activation by some serine proteases, arginine esterases in rat submandibular gland. Biochemical and Biophysical Research Communications, 1990, Volume 166, Issue 1, Pages 231-237
- Jonas Anders, Michael Kemme. The binding of tissue prokallikrein to isolated human neutrophils. FEBS Letters, 1994, Volume 348, Issue 2, Pages 166-168
- Manfred Maier, Ingrid Jerabek, Günther Reissert, Eva Höltzl, Bernd R. Binder. Correlation of two different assays for urinary kallikrein in normotensive and hypertensive subjects. Clinica Chimica Acta, 1988, Volume 178, Issue 2, Pages 127-139
- Bruce L. Zuraw, Sandra Sugimoto, C. Lowell Parsons, Tony Hugli, et al. Activation of Urinary Kallikrein in Patients with Interstitial Cystitis. The Journal of Urology, 1994, Volume 152, Issue 3, Pages 874-878
- Claudio Ferri, Cesare Bellini, Antonio Carlomagno, Alessandro Perrone, Anna Santucci. Urinary kallikrein and salt sensitivity in essential hypertensive males. Kidney International, 1994, Volume 46, Issue 3, Pages 780-788
- Ayad A. Jaffa, Donald H. Miller, Ricardo H. Silva, Harry S. Margolius, Ronald K. Mayfield. Regulation of renal kallikrein synthesis and activation by glucocorticoids. Kidney International, 1990,Volume 38, Issue 2, Pages 212-218
- Julie Chao, Lee Chao. Rat urinary kallikrein. Methods in Enzymology, 1988, Volume 163, Pages 128-143
- Gary P Richards, Lee Chao, Julie Chao. Distribution of Tissue Kallikreins in Lower Vertebrates: Potential Physiological Roles for Fish Kallikreins. Comparative Biochemistry and Physiology Part C: Pharmacology, Toxicology and Endocrinology, 1997, Volume 118, Issue 1, Pages 49-58
- KazuyukiKizuki,TomohikoSuzuki,Motoshige Kudo, Tetsuya Noguchi. Immunohistochemical demonstration of tissue kallikrein in the neurons of rat brain. Brain Research, 1994, Volume 634, Issue 2, Pages 305-309
- Gary P. Richards, Julie Chao, Lee Chao. Tissue kallikreins in evolutionarily diverse vertebrates. Immunopharmacology, 1996, Volume 32, Issues 1-3, Pages 94-95
- Reinhard Geiger, Hans Fritz. Human urinary kallikrein. Methods in Enzymology, 1981, Volume 80, Pages 466-492
- Masanori Takaoka, Hirofumi Okamura, Yoshikazu Kuribayashi, Hidehito Matsuoka, Shiro Morimoto. Activation of urinary inactive kallikrein by an extract from the rat kidney cortex. Life Sciences, 1985,Volume 37, Issue 11, Pages 1015-1022
- Gasteiger E, Hoogland C, Gattiker A, et al. Protein Identification and Analysis Tools on the ExPASy Server, Proteomic Protocols Handbook, 2005, Volume 112, Pages 571-607
- C Geourjon, G Deléage. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences Cabios, 1995, Volume 11, Issues 6, Pages 681-684
- Nikolaj Blom, Thomas Sicheritz-Pontén, Ramneek Gupta, et al. Kinase specific predictions: Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 2004, Volume 4, Issues 6, Pages 1633-1649
- Gasteiger E, Hoogland C, Gattiker A, et al. Protein Identification and Analysis Tools on the ExPASy Server, Proteomic Protocols Handbook, 2005, Volume 112, Pages 571-607