Information
Journal Policies
Quantification of DNA Extracted from Libyan Buccal Swabs by Using Equation of Standard Curve
Samir Elmrghni1*
Copyright :© 2018 Authors. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Since DNA is the blueprint for life, everything living contains DNA. DNA isolation is one of the most basic and essential techniques in the study of DNA. The extraction of DNA from cells and its purification are of primary importance to the field of biotechnology and forensics. Extraction and purification of DNA are the first steps in the analysis and manipulation of DNA that allow scientists to detect genetic disorders, produce DNA fingerprints of individuals, and even create genetically engineered organisms that can produce beneficial products such as insulin, antibiotics, and hormones. DNA can be extracted from many types of cells. The standard curve Calibration was used to calculate the DNA quantities extracted from bucal swabs in samples From Benghazi population which included in different research studies.
Keywords: standard curve, DNA quantification, Libyan,Forensic Science
1.Introduction
Libya, a Northern African country, was first inhabited by Berbers, followed by Phoenicians, Greeks, Romans, Arabs and Ottomans. Libya became independent in 1951 after a brief period as an Italian colony; it had been invaded by Italy in 1911. In February 2011 an uprising against the Libyan government occurred. Benghazi is the second largest city in Libya and the main city (or capital) of the Cyrenaica region (or exProvince), located in the North of Africa. Benghazi is located half way between Tripoli in the West (a distance of approximately 1000 Km between these cities) and Cairo in the East (also approximately 1000 Km). Cyrenaica is surrounded by desert on three sides; hence in ancient times the most accessible civilization was to the North, across the Mediterranean, in Crete and Greece, only 400 km away.
The population of Benghazi was 500,120 in 1995 (census) and increased to 670,797 in the 2006 census. As with other cities in Libya, there is a reasonable amount of ethnic diversity in Benghazi. The people of eastern Libya, Benghazi included, have in the past always been of predominantly Arab descent. In recent times, however, there has been an influx of African immigrants into Benghazi. There are also many Egyptian immigrants in Benghazi and a small Greek community also exists in Benghazi; the Greek island of Crete is a short distance away from Benghazi and many families in Benghazi today bear Cretian surnames. In modern times, Benghazi has seen a lot of Libyans from different parts of the country move into the city, especially since the Kingdom era (1951-1969). Many Libyans came to Benghazi from Misrata (About 60% of the population have roots from Misrata, West of Benghazi).
2. Quality Control
The laboratory has participated in the Y-STR Haplotyping Quality Assurance Exercise (Certified at 2010-5-20). The data were submitted to YHRD (www.yhrd.org) and received the accession number: YA003680 DNA was quantified using a method developed in the laboratory. Lambda DNA (New England BioLabs, Hitchin, UK) was serial diluted to prepare standard solutions with known quantities of DNA in 10μl volumes: 200 ng, 100 ng, 20 ng, 2 ng, 0.2 ng and 0.02 ng. The 10 μl volumes of the DNA standard solutions and 10μl volumes of volunteer samples were added in triplicate to a Fast Optical 96-well reaction plate (Applied Biosystems). A volume of 10 μl SYBR® Green I nucleic acid gel stain 10,000x.
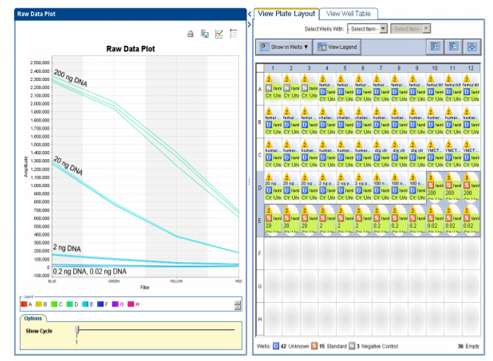
(SIGMA) diluted to 20x concentrated was then added to each well. The plate was then centrifuged at 3,000 rpm for 1 min and maintained at room temperature for 5 min before reading (protected from light). The plate was read using a StepOnePlus™ Real-Time PCR System (Applied Biosystems) under the following conditions: initial holding of 30 seconds at 25ºC and 1 cycle of 10 seconds at 25°C with data collection. The raw data was then analysed as shown in table below. A screen capture print of the raw data obtained for the standard DNA quantities is shown in the figure 1 below.
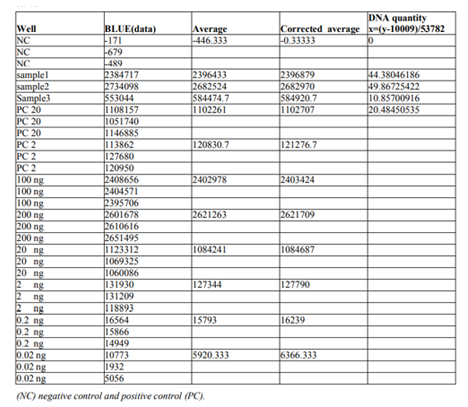
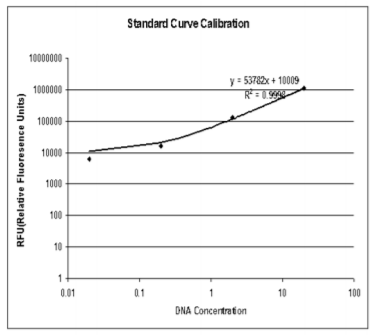
The raw data was then exported as an Excel file and a standard curve drawn (with logarithmic scale for the X and Y axis and adding a trendline) using the blue amplitude values Table 1. (RFU) obtained for the DNA quantities 20 ng, 2 ng and 0.2 ng (200 ng and 0.02 ng not included due to saturation and below threshold, respectively) Table 1. The standard curve is shown in the figure3 below.
The standard curve Calibration was then used to calculate the DNA quantities in the unknown samples ( the ideal concentration used to type Autosomal and Y- filer kits is 0.1ng) by substituting Y for the blue amplitude values. DNA concentrations were then calculated noting that 10 μl had been applied to each well. The values obtained for the positive controls (20 ng and 2 ng) indicated the accuracy of the equation.
For Benghazi population samples which included in different research studies (1, 2, 3&4) it was discovered that a 10 fold dilution of the samples gave an optimum amount of target DNA in the amplifications.
References
- Samir Elmrghni, Ron A Dixon, Yvette M Coulson-Thomas and D. Ross Williams. (2011). Population genetic data for 17 Y STR markers from Benghazi (East Libya), Forensic Sci. Int. Genet. doi:10.1016/j.fsigen. 2011.05.001.
- Samir Elmrghni, S, Yvette M Coulson-Thomas, Mahmoud Kaddura, Ron A Dixon and D. Ross Williams. (2011). Genetic data provided by 15 autosomal STR loci in the Libyan population living in Benghazi, Forensic Sci. Int. Genet. doi:10.1016/j.fsigen.2011.07.006.
- Samir Elmrghni, Ron A Dixon and Ross Williams (2011) Frequencies of C282Y and H63D mutations in the haemochromatosis (HFE) gene in Libyans living in Benghazi. Int J ClinExp Med 2010;4(3):200-204.
- Samir Elmrghni, Ron A Dixon and Ross Williams (2012)Controversial cases of human gender identification by amelogenin test in some Libyans (Benghazi City) (International Indian Journal of Forensic Medicine and Toxicology).;July-Dec.,2012, Vol.6,No.2