Information
Journal Policies
Implication of Neighbors in the Genetic Diversity of the Human Immunodeficiency Virus Type 1 in the Democratic Republic of Congo
Erick Ntambwe Kamangu*
Copyright : © 2018 . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The distribution of HIV-1 in the Democratic Republic of Congo (DRC) is very heterogeneous; it is very dynamic, evolving and unpredictable. The DRC has the largest number of variants of HIV type 1, particularly Group M. A research on the published works and the abstracts presented in conference having as subject of interest the identification of the different variants of HIV type 1 in DRC from 1997 to 2015 was done. The research of these works published on the different strains was made on the Internet on the research websites. According to the articles and abstracts of conferences published since 1997, a dominant prevalence of group M (100%) has been noted. For the whole country, the strains are found in the following order: A (49.40%), G (10.73%), C (9.01%) and D (7.86%). The geographical distribution of HIV-1 variants in the DRC is closely related to the distribution of variants in neighboring countries. This distribution is a true mosaic; it is different according to the provinces, the geographical distribution in the country and the methods used.
Variants HIV-1, DRC, Neighboring Countries, AIDS
The distribution of Human Immunodeficiency Virus Type 1 (HIV-1) in the Democratic Republic of Congo (DRC) is very heterogeneous; it is very dynamic, evolving and unpredictable [1]. Located in central African region, the DRC has the largest number of variants of HIV type 1, particularly of Group M in terms of subtypes, sub-subtypes and CRFs circulating across the country [2,4-6].
A research on the published works and the abstracts presented in conference having as subject of interest the identification of the different variants of HIV type 1 in DRC from 1997 to 2015 was done. The search for these published works on the different strains was done on the internet on research websites. It was based on the following keywords: "HIV, subtype, DRC", "genotype, HIV, DRC" and "HIV strains in the DRC".
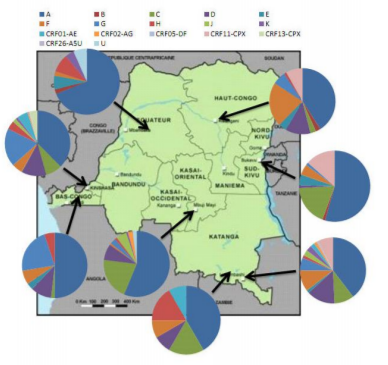
According to articles and conference abstracts published from 1997 to 2015, HIV-1 group M is 100% dominant and subgroup A is almost 50% [31,2-68,9] overall in the DRC. The distribution of subtypes differs from one part of the country to another as shown in the figure (1) below. In the Eastern part of the country, subtype a (44.73%) is dominant on subtypes C (12.20%), G (11.5%), D (9.12%) and U (7.24%). In the center part, subtypeA (62.57%) is followed by subtypes C (10.32%), H (5.02%), U (4.3%) and D (3.9%). In the Western part, subtypeA (40.91%) is followed by subtypes G (19.29%), D (10.5%), F (5.65%) and C (4.51%). For the country, the subtypes are found in the following order: A (49.40%), G (10.73%), C (9.01%) and D (7.86%) [7]. Inter and intra-group differences are statistically significant for the different strains (p> 0.00).
The geographical distribution of HIV-1 variants in the DRC is closely related to the distribution of variants in neighboring countries [8]. Among the northeastern neighbors (Sudan[9, Uganda [3,10,11], Rwanda [3,12], Burundi [3,13,14]), the predominant subtypes are D, C, A and B. While among the Southeast neighbors (Tanzania [3,15], Zambia [3,16] and even Kenya [3,16,17]), the dominant subtypes are A, C and D. This would explain the difference in prevalence in the eastern part of the country between Kisangani and Bukavu in the northeast, and Lubumbashi and Likasi in the southeast. The neighbor of the South, Angola [3,18], has a dominance of subtypes A, C and H which is reflected on Kinshasa and Kimpese. The neighbor of the West, the Republic of Congo [3,19], has a dominance of subtypes A, G and D which is also reflected on Kinshasa and Kimpese. The neighbor of North, the Central African Republic [3,20], has a dominance of subtypes A, B, D and CRF01_AE which is reflected on Bwamanda. The rural exodus to big cities such as Kinshasa, Lubumbashi and Mbuji Mayi, the displacement of the victims of the wars in the East of the country as well as the movement of the populations across the borders of the DRC make this epidemic difficult to control for the country.
The distribution of the different variants of HIV Type 1 in the DRC is a mosaic; it is different according to the provinces and the geographical distribution in the country. This diversity will quickly become a big problem for the fight against HIV in the DRC if it is not under control.
References
- Peeters M, Mulanga-Kabeya C et Delaporte E : La diversité génétique du VIH Type 1. Virolog ie 2000 ; 4 (5) : 371-881
- Etienne L et Peeters M : Origine du VIH, une réussite émergentielle. Virologie 2010 ; 14 (3) :171-83
- Papathanasopoulos MA, Hunt GM and Tiemessen CT: Evolution and Diversity of HIV-1 in Africa: a Review. Virus Genes. 2003; 26 (2): 151-63
- Janssens W, Buvé A and Nkengasong JN: The Puzzle of HIV-1 Subtypes in Africa. AIDS 1997; 11: 705-12
- Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JMM, Kalengayi RM, Marck EV, Gilbert MTP and Wolinsky SM: Identification of a Complex env Subtype E HIV Type 1 Virus from the Democratic Republic of Congo, Recombinant with A, G, H, J, K, and Unknown Subtypes. Nature 2008; 455: 661-4
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B and Delaporte E: Unprecedented degree of Humain Immunode ficiency Virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggest that the HIV-1 pandemic originated in Central Africa. J Virol. 2000; 74: 10498-507
- Kamangu NE, Kabututu Z, Mvumbi LG, Kalala LR, Mesia KG. Genetic Diversity of Human Immunodeficiency Virus Type 1 in the Democratic Republic of Congo: a review of available data. International Journal of Collaborative Research on Internal Medicine & Public Health. 2013; 5 (5): 295-309
- Vidal N, MulangaC, EdidiBazepeo S, Mwamba J, Tshimpaka JW, Kashi M, Mama N, Laurent C, Lepira F, Delaporte E and PeetersM : Distribution of HIV-1 Variants in the Democratic Republic of Congo Suggests Increase of Subtype C in Kinshasa Between 1997 and 2002. J AIDS. 2005; 40 (4): 456-62
- Hierholzer M, Graham RR, El Khidir I, Tasker S, Darwish M, Chapman GD, Fagbami AH, Soliman A, Birx DL, McCutchan F and Carr JK: HIV Type 1 Strains from East and West Africe are Intermixed in Sudan. AIDS Res Hum Retroviruses. 2002; 18 (15): 1163-6
- Hu DJ, Baggs J, Downing RG, Pieniazek D, Dorn J, Fridlund C, Biryahwaho B, Sempala SDK, Rayfield MA, Dondero TJ and Lal R: Predominance of HIV-1 Subtype A and D Infections in Uganda. Emerging Infectious Diseases. 2000; 6 (6): 609-15
- Collinson-Streng AN, Redd AD, Sewankambo NK, Serwadda D, Rezapour M, Lamers SL, Gray RH, Wawer MJ, Quinn TC and Laeyendecker O: Geographic HIV Type 1 Subtype Distribution in Rakai District, Uganda. AIDS Res Hum Retroviruses. 2009; 25 (10): 1045-48
- Roman F, Karita E, Monnet A, Lambert C, Fontaine E, Allen S, Schneider F, Hemmer R, Schmit JC and Arendt V : Rare and new V3 loop variants in HIV-1 positive long-term non progressors from Rwanda. AIDS 2002; 16: 1827-9
- Koch N, Ndihokubwayo JB, Yahi N, Tourres C, Fantini J and Tamalet C: Genetic Analysis of HIV Type 1 Strains in Bujumbura (Burundi): Predominance of Subtype C Variant. AIDS Res Hum Retroviruses. 2001; 17 (3): 269-73
- Vidal N, Niyongabo T, Nduwimana J, Butel C, Ndayiragije A, Wakana J, Nduwimana M, Delaporte E and Peeters M: HIV Type 1 Diversity and Antiretroviral Drug Resistance Mutations in Burundi. AIDS Res Hum Retroviruses. 2007; 23 (1): 175-80
- Mosha F, Urassa W, Aboud S, Lyamuya E, Sandstrom E, Bredell H and Williamson C: Prealence of Genotypic Resistance to Antiretroviral Drugs in Treatment-Naïve Youths Infected with Diverse HIV Type 1 Subtypes and Recombinant Forms in Dar es Salaam, Tanzania. AIDS Res Hum Retroviruses. 2011; 27 (4): 377-382
- Morison L, Buvé A, Zekeng L, Heyndrickx L, Anagonou S, Musonda R, Kahindo M, Weiss HA, Hayes RJ, Laga M, Janssens W and Van der Groen G, for the Study Group on Heterogeneity of HIV Epidemics in African Cities: HIV-1 Subtypes and HIV Epidemics in four Cities in Sub-Saharan Africa. AIDS 2001; 15 (suppl 4): S109-16
- Yang C, Li M, Shi YP, Winter J, Van Eijk AM, Ayisi J, Hu DJ, Steketee R, Nahlen B and Lal RB: Genetic Diversity and High Proportion of Intersubtype Recombinants among HIV Type 1 Infected Pregnant Women in Kisumu, Western Kenya. AIDS Res Hum Retroviruses. 2004; 20 (5): 565-74
- Abecasis A, Paraskevis D, Epalanga M, Fonseca M, Burity F, Bartolomeu J, Carvalho AP, Gomes P, Vandamme AM and Camacho R : HIV-1 Genetic Variants Circulation in the North of Angola. Infection, Genetics and Evolution. 2005; 5: 231-37
- Candotti D, Tareau C, Barin F, Joberty C, Rosenheim M, M’pele P, Huraux JM and Agut H: Genetic Subtyping and V3 Serotyping of Type 1 Isolates in Congo. AIDS Res Hum Retroviruses 1999; 15 (3): 309-14
- McCutchan FE: Understanding the genetic diversity of HIV-1. AIDS 2000; 14Suppl 3: S31-44