Information
Journal Policies
Increase of Recombinant forms CRF20, 23, 24_BG and Several URF of HIV-1 among Newly Diagnosed Cuban Patients: 2013-2014
Liuber Y Machado1,Yanet Pintos2,Hector M Diaz3,Lissette Perez4,Madeline Blanco5,Vivian Kouri7,Yoan Aleman8,Liodelvio Martinez9,Marta Dubeda10,Carlos Aragones11,Nancy Ruiz12,Eladio Silva13,Yudira Soto14,Neisy Valdes15,Yoanna Banos16,YanirisCaturla17,Dania Romay18,Jorge Perez19,Carmen Nibot20,Niurka Rocha21,Rene Rodriguez22,Maria L Sanchez23,Aldo Trinquete24
2.Instituto de Medicina Tropical Pedro Kourí, Havana, Cuba.
3.Hospital Clínico Quirúrgico Hermanos Ameijeiras, Havana, Cuba.
4.HIV/AIDS Program, Ministry of Public Health, Cuba The fours first authors contributed equally to this study.
Copyright : © 2017 . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The objective of this study was characterize the genetic diversity of HIV-1 in Cuban patients diagnosed during the period 2013-2014. A total of 189 patient samples from all provinces of the country, diagnosed withHIV-1 infection, were studied during the period from April 2013 to April 2014. Viral RNA was isolated for further amplification of the pol gen of HIV-1. The purified products were sequenced and the viral subtype was determined. The recombination was evaluated by boots canning. The relationship between viral variants and epidemiological, immunological and virological variables was determined. 27.5% of the samples were classified as CRF20, 23, 24_BG, followed by subtype B (23.8%) and the presence of other recombinants as CRF19_cpx and CRF18_cpx. 11.1% of the samples were classified as URF and the most frequent combination detected was between CRF18_cpx and CRF19_cpx. The CRF19_cpx variant was detected more frequently in Havana. There was no association between viral variants and sexual orientation. The study confirms the high genetic diversity of HIV-1 in the Cuban population diagnosed between 2013 and 2014 and the predominance, for the first time of CRF20, 23, 24_BG over subtype B in the sample studied, which supports the importance of maintaining close epidemiological surveillance.
molecular epidemiology, HIV-1, subtype, circulating recombinant forms, unique recombinant ,AIDS
1. Introduction
The origin of Human Immunodeficiency Virus type 1 (HIV-1) has been associated with several events of zoonotic transmission of the Simian Immunodeficiency Virus (SIV) of chimpanzees to humans in Central and West Africa, occurring at the beginning of the century XX [1]. From the phylogenetic point of view, HIV-1 is classified into four groups: M, N, O and P [2]. Group M, responsible for the majority of HIV-1 infections in the world, initially spread throughout Africa [3] and in response to various genetic forces was diversified into different subtypes, sub-subtypes, more than 70 circulating recombinant forms (CRF) and multiple unique recombinant forms (URF) [4]. It has been suggested that the presence of circulating recombinant types, subtypes and forms of HIV in a specific geographical area, leads to generations of recombinant forms that emerge in individuals with dual or multiple infections [5].
The HIV epidemic in Cuba began in 1986 and since its inception, the strategies implemented by the Ministry of Public Health have made possible to detect the circulation of HIV-1 and HIV-2 in the HIV-positive population [6]. The first molecular epidemiological studies in Cuba described the presence of several HIV-1 subtypes, with a higher frequency in subtype B and the presence of non-B subtypes introduced from the African continent [7]. Subsequent studies detected mosaics of genetic variants in the HIV-1 genome in samples from Cuban patients and described the first recombinants in the epidemic context [8,9]. The introduction of antiretroviral therapy in Cuba in 2001 led to the monitoring of HIV-1 resistance to antiretrovirals in treated and untreated patients, and in addition to providing the resistance profile to the different antiretrovirals, allowed us to know the Viral subtype that was infecting the individuals [10-12]. One of the objectives of epidemiological surveillance is to know the circulating genetic variants of HIV-1 in the newly diagnosed population, due to the possible implications of genetic variability in diagnosis, transmissibility, clinical progression and antiretroviral therapy. These elements motivated the objective of the present study: To characterize the genetic diversity of HIV-1 in Cuban individuals newly diagnosed during the period 2013-2014.
2. Materials And Methods
A cross-sectional analytical study was performed on a representative sample of all patients diagnosed withHIV-1 infection during the period from April 2013 to April 2014. A randomized stratified probabilistic sampling was performed, with a prevalence of 25 % and a 5% of error of the sample. All provinces of the country were represented and 189 patients with HIV-1 viral load values over 1000 HIV-1 RNA copies / mL were studied.
Ethical procedures were carried out in accordance with the requirements or standards of the Ministry of Public Health of the Republic of Cuba and the Ministry of Science, Technology and Environment (CITMA), which contemplates the principles laid down in the Declaration of Helsinki for medical research in Human beings and the LISIDA and the IPK Ethics Committees. Prior to sample collection, informed consent was obtained from each patient who participated in the study.
The CD4 + cell count was determined using Becton Dickinson technology (Biosciences,USA). Plasma viral load was determined using COBAS Ampliprep / COBAS Taqman HIV-1 Test (Roche Diagnostics GmbH, Mannheim,Germany).
Extraction of viral RNA, amplification of the protease and RT regions of the HIV-1 pol gene by Reverse Transcriptase-nested PCR and subsequent nucleotide sequencing were performed according to the procedures described by Alemán et al, 2015 [13]. The HIV-1 subtype was determined by the REGA HIV-1 subtypingtool v 3.0 program (http://dbpartner s.stanford.edu:8080/RegaS ubtyping/stanford-hiv/typingtool/) and was confirmed by phylogenetic analysis. To corroborate the results obtained, the nucleotide sequences of the 189 samples studied were automatically aligned with reference sequences obtained from the HIV Database of the National Laboratory of Los Alamos (www.hiv-web.lanl.gov) using the tool Clustal W of the MEGA program package version 6.0 [14]. For the determination of possible recombinants and breakpoints, boot scaning was performed and the points of similarity between sequences were determined using the Simplot v 3.5 [15] and RDP4 [16] programs. Aditionally, phylogenetic tree was constructed using the neighbor joining method and the genetic distance was estimated according to the two parameters of Kimura. Bootstrap values were calculated based on 1000 replicates. The tree was visualized using the program Fig Tree v 1.1.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Mean and standard deviation (SD), median and interquartile range (IQR), and frequencies (%) were used to describe patient characteristics. The Χ2 test and the Fisher exact test were used to compare categorical variables and continuous variables. The non parametric Kruskal Wallis test was used for the comparison of medians of categorical variables. The variables that presented p< 0.05 were considered in a multivariate logistic regression model after evaluating the multicollinearity of the variance inflation factors. The selected variables were included in a multiple logistic regression model with a significance level of (p < 0.05). Odds ratios (ORs) were estimated with a 95% confidence interval (CI). A value of p < 0.05 was considered statistically significant. All statistical analyzes were performed using the SPSS 18 statistical package (SPSS Inc., Chicago, IL).
3. Results
The present study included 189 patients diagnosed with HIV-1 in the period between April 2013 and April 2014. The mean age of the patients studied was 33 years with values ranging from 17 to 74 years? 80.9% of the patients were male and 80.3% were in the risk behavior group of men who have sex with other men (MSM). The province of Havana was the region of the country that provided the largest number of patients for the study (38.6%). The median viral load was 58,000 copies of RNA / mL and the CD4 + cell count was 371 cells / mm3 (Table 1).
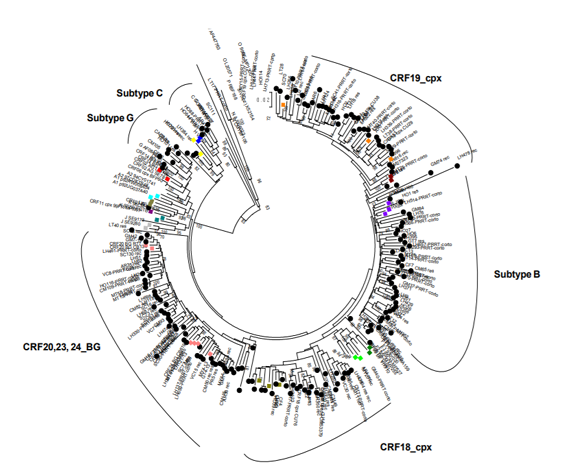
In the present study the viral variants CRF20, 23, 24_BG (27.5%) were the most frequently observed, followed by subtype B (23.8%). However, other subtypes were also detected: G (4.2%), C (2.1%), H (1.05%) and circulating recombinant forms such as CRF19_cpx (19.5%), CRF18_cpx (9.5%) and CRF01_AE (0.5 %). In the phylogenetic tree the clusteringof these viruses with their respective reference sequences is observed In contrast, 11.1% of the samples were not grouped with reference sequences according to phylogenetic criteria and they were classified as URF(Figure. 1).
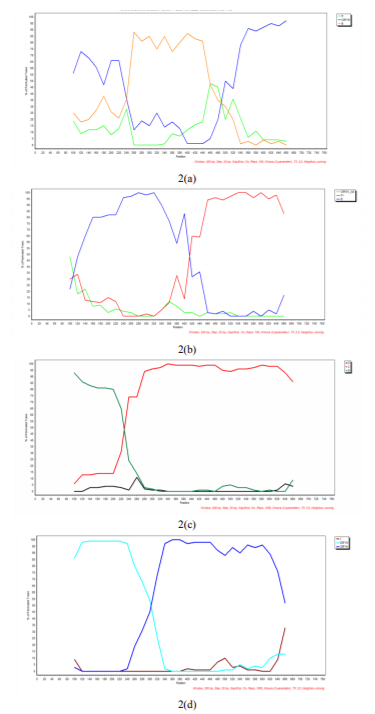
The recombination analysis detected as more frequent URF combinations: CRF19_cpx/B, B/F1, B/C and CRF18_cpx/CRF19_cpx, as illustrated in Figure 2.
The MSM population of the present study was more frequently infected with subtype B (31/123, 25.2%), followed by CRF20, 23, 24_BG (28/123, 22.8%), CRF19_cpx (23 / 123, 18.6%), URF (15/123, 12.1%), and CRF18_cpx (13/123, 10.5%). However, the same proportion of patients infected with CRF20, 23, 24_BG, in both MSM and HT (53.8% (28/52) and 41.6% (24/52), respectively) is observed. Subtype B was associated with the age range of 46-61 years (p = 0.019, OR = 2844 (1.262-6.406)), and the CRF20, 23, 24_BG variants with the ranges of 15 to 30 years (p = 0.035, OR = 2,053 (1,069-3,944)) (Table 2). Linear regression analysis showed that CRF19_cpx was two times more frequently detected amongpatients living in Havana (p = 0.039, OR = 2.198 (1.062-4.549)) while it was three times more likely to detect CRF18_cpx in patients residing in the central region than in the rest of the country (p = 0.017, OR = 3,716 (1,316-10,496)). URF was associated with CD4 + cell counts below 200 cells (p = 0.050, OR = 3.152 (1.150-8.640)) (Table 2). No significant differences were found when comparing medians of viral load values between the different subtypes using Kruskal Wallis non-parametric test (results not shown).
4. Discussion
Since the detection of the first case of HIV in the 1980s, the genetic diversity of this retrovirus has played a fundamental role in its dissemination, consequently, epidemiological surveillance of circulating viral variants is a fundamental premise for better management and control of the epidemic. At present, the scientific community has givenspecial attention to the non-B subtypes, due to the increase of these variants in areas such as North America and Europe, where previously subtype B prevailed. This could have implications in the response to antiretroviral therapy, progression of the disease and the design of vaccines . Although Cuba has a low prevalence of HIV infection (0.19%), several studies have described a high genetic diversity of HIV-1, rendering into a wide circulation of subtypes, sub-subtypes, CRFs and multiple URFs (7 -12), comparable to the variability reported in some African countries [4]. Contrary to what has been described so far in the Cuban epidemic, where subtype B has been the most frequent genetic form, in the present study CRF20, 23, 24_BG were the predominant genetic variants (28%) in the studied population. Previous studies described a decrease in subtype B with an increase in non-B subtypes and several recombinants in samples collected in 2003 [9]. The increase in the frequency of detection of CRF20, 23, 24_BG in several studies carried out in the newly diagnosed population from 2009 to the present, reinforces the fact of the displacement of subtype B by these recombinant forms [11,12]. Similar events are reported in some localities of Brazil with the recombinant BF and BC [17], as well as in Russia with the recombinant AB [18] and in China with the recombinant BC [19,20], mainly in intravenous drug users IDU). It is not known exactly whether the rapid expansion of BG recombinants is due to viral biological characteristics or increased risk sexual behaviors, or both, but such diversity reflects the dynamic nature of the genetics of the HIV-1 epidemic, through which new genetic forms, introduced or generated by recombination, can rapidly expand into an established epidemic [9].
Interestingly, 11.1% of the viral infections of the present study were caused by URFs. These recombinants are very common in regions where multiple subtypes circulate, such as sub-Saharan Africa; But recentlyit has been detected relatively frequently in developed countries [21]. Despite the spread of recombinants in the HIV-1 pandemic, the time of origin is not well known. It has been suggested that early recombination was a common phenomenon in the historical evolution of HIV-1. However, the detection of viral URFs could increase in the global epidemic context if more regions of the viral genome were analyzed. Recombination seems to be very important in the evolution of HIV-1, since it can provide a biological advantage over parental viruses, facilitate biological adaptation and increase evolutionary fitness or viral capability. In addition, it has complex consequences on the estimation of the time of divergence, due to an apparent increase in the ranges of variation between the nucleotide sites and because it reduces the genetic distance between the sequences [21]. However, the prevalence of URF at the global level (estimated at around 20%) is still underestimated. Genetic complexity is not always detected, and this is mainly due to the subtyping of only one genetic region and not the complete viral genome [4]. The MSM population of the present study was more frequently infected insubtype B (25.2%). Subtype B has been associated with MSM in the Cuban epidemic and worldwide [9,23,24]. Statistical analysis of the present study showed no association between sexual orientation with this subtype, nor with the rest of the genetic forms detected (Table 2), a result that differs from that described by previous studies conducted in Cuban patients infected with HIV-1 [7,9].
The most frequent detection of CRF19_cpx in Havana and CRF18_cpx in the central region reflect the dynamics of this retrovirus in the Cuban epidemic context. In a study carried out by Perez et al., 2007, patients from the central region of the country were associated with CRF19_cpx, while URF CRF18_cpx, B / CRF18_cpx and subtype H were associated with patients residing in the eastern region [25]. The discrepancy between these results could be explained by the time elapsed between the two studies, the diversity of genetic forms that circulate in the Cuban epidemic, which, together with the migration of infected individuals from one region to another, has made possible an increase in some viral variants in regions where their proportion was lower.
The detection of URF was associated with CD4 + cell counts below 200 cells; interestingly10 of these viruses contained the combinations ofCRF19_cpx / CRF18_cpx. It has been argued that the genetic diversity of HIV-1 might affect the rate of progression to AIDS. However, several authors associate subtypes C, and D with a rapid progression of the disease and categorize themas more aggressive than the G, CRF01_AE variants,CRF02_AG and A, being subtype A considered as the less aggressive HIV-1 subtype. One previous study showed the association of CRF19_cpx with the rapid progression to AIDS in Cuban patients [27]. Therefore, continuous virological, immunol - ogical and clinical monitoring of individuals infected with this recombinant or presenting this variant in the genome architecture viral is recommended.
5. Conclusions
In conclusion, this study confirms the high genetic variability of HIV-1 in a group of patient samples diagnosed between April 2013 and April 2014. Analysis of the pol gene in our study allowed to verify the frequency and diversity of recombinant forms, and the displacement of subtype B by CRF20, 23, 24_BG as a predominant variant in the studied population, in comparison to previous studies carried out in Cuba. The results obtained in addition to updating the epidemiological situation to the surveillance program of the Cuban Ministry of Public Health, would reinforce studies on the implications of the genetic diversity of HIV in the epidemic, pathogenesis, vaccine development and the design of new therapeutic strategies.
References
- Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 455: 661-664.(2008)
- Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Phil. Trans. R. Soc. B. 365: 2487-2494.(2010)
- Faria NR, Rambaut A, Suchard M, Baele G, Trevor B, Ward MJ, et al. The early spread and epidemic ignition of HIV-1 in human populations. Science. 346 (56). Doi:10.1126/science.1256739. (2014)
- Santoro MM, Perno CF. HIV-1 genetic variability and clinical implications. ISRN Microbiology article ID 481314: 20 pages. (2013)
- Allen TM, Altfeld M. HIV-1 super infection. J. Allergy ClinImmunol. 112: 829-35. (2003)
- Díaz HM, Pérez MT, Lubián AL, Nibot C, Cruz O, Silva E, et al. HIV Detection in Cuba: Role and Results of the National Laboratory Network. MEDICC Review. 13(2): 9-13. (2011)
- Rolo F, Miranda L, Wainberg M, Gu Z, Lobaina L, Noa E, et al. Envelope V3 Region Sequences of Cuban HIV-1 Isolates. J Acquir Immune DeficSyndr and Hum Retrovirology. 9:123-125. (1995)
- Cuevas MT, Ruibal I, Villahermosa ML, Díaz H, Delgado E, Parga EV, et al. High HIV-1 genetic diversity in Cuba. AIDS. 16:1643-1653. (2002)
- Pérez L, Thomson M, Aragonés C, González Z, Pérez J, Casado G, et al. HIV Type 1 Molecular Epidemiology in Cuba: High Genetic Diversity, Frequent Mosaicism, and Recent Expansion of BG Intersubtype Recombinant Forms. AIDS Res Hum Retroviruses. 22 (8) : 724-733. (2006)
- Kourí V, Alemán Y, Pérez L, Pérez J, Fonseca C, Correa C, et al. High frequency of antiviral drug resistance and non-B subtypes in HIV-1 patients failing antiviral therapy in Cuba. J ClinVirol. 55: 348-355. (2012)
- Machado LY, Blanco M, Dubed M, Díaz HM, Ruiz NM, Valdés N, et al. HIV type 1 genetic diversity in newly diagnosed Cuban patients. AIDS Res HumRetroviruses. 28 (8): 956-960. (2012)
- Pérez L, Kouri V, Alemán Y, Abrahantes Y, Correa C, Aragonés C, et al. Antiretroviral drugresistance in HIV-1 therapy-naivepatients in Cuba. Infect Genet Evol. 16C: 144–150. (2013)
- Alemán Y, Vinken L, Kourí V, Pérez L, Álvarez A, Abrahantes Y, et al. Performance of an in house Human Immunodeficiency Virus type 1 genotyping system for assessment of drug resistance in Cuba. PLoS ONE. 10 (2): e0117176.Doi:10.1371/journal.pone. 0117176. (2015)
- Tamura KD, Peterson D. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10): 2731-39. (2011)
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 73 (1): 152-60. (1999)
- Martin DP, Murell B, Golden M, Khoosal A and Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution. 1 (1): vev003. (2015)
- da Costa Carolina Marinho, Costa de Oliveira Cintia Mara, Chehuan de Melo Yonne Francis, Delatorre Edson, Bello Gonzalo, and Couto-FernandezJose Carlos. High HIV-1 Genetic Diversity in Patients from Northern Brazil. AIDS Res Hum Retroviruses. 32(9): 918-922. doi:10.1089/aid.2016.0044. (2016)
- Gashikova NM, Astakhova EM, Gashinikova MP, Bocharov EF, Petrova SV, Pun´ko OA et al. HIV-1 epidemiology, genetic diversity, and primary drug resistance in the Tyumen Oblast, Russia. BioMed Research International. http://dx.doi.org/10.1155/2016/2496280. (2016)
- Li Xiaoshan, Li Wei, Zhong Ping, Fang Kun, Zhu Kexin, Musa Taha Hussein, Song Yue, Du Guoping, GaoRong, Guo Yan, Yan Wenjuan, Xuan Yang, and Wei Pingmin. Nationwide Trends in Molecular Epidemiology of HIV-1 in China.AIDS Res Hum Retroviruses. 32(9): 851-859. (2016)
- Chen Xin, Ye Mei, Pang Wei, Smith Davey M., Zhang Chiyu, and Zheng Yong-Tang. First Appearance of HIV-1 CRF07_BC and CRF08_BC Outside China. AIDS Res Hum Retroviruses. 33(1): 74-76. doi:10.1089/aid.2016.0169. (2017)
- Yebra G, Holguín A, Pillay D, Hue S. Phylogenetic and demographic characterization of HIV-1 transmission in Madrid, Spain. Inf Gen Evol. 14: 232-239. (2013)
- Vuilleumier S, Bonhoeffer S. Contribution of recombination to the evolutionary history of HIV. CurrOpin HIV AIDS. 10(2):84-9. (2015)
- Avila MM, Pando MA, Carrión G, Peralta LM, Salomón H, Carrillo MG, et al. Two HIV-1 epidemics in Argentina: different genetic subtypes associated with different risk groups. J Acquir Immune DeficSyndr. 29 (4): 422-6. (2002)
- Blanco M, Machado LY, Díaz HM, Ruiz N, Romay D, Silva E. HIV-1 genetic variability in Cuba and implications for transmission and clinical progression. MEDICC Review. 17 (4): 25-31. (2015)
- Pérez L, Pérez Alvarez L. Carmona R, Aragonés C, Delgado E, Thomson M, et al. GenotypicResistenceto Antiretroviral drug in patients infected with several HIV type I genetic forms in Cuba. AIDS Res Hum Retroviruses. 23 (3): 407-414. (2007)
- LeiteThaysse Cristina Neiva Ferreira, Campos Dayse Pereira, Coelho AlessandraBrum, Teixeira Sylvia LopesMaia, VelosValdilea,MorgadoMarizaGonçalves, and GuimarãesMonickLindenmeyer. Impact of HIV-1 Subtypes on AIDS Progression in a Brazilian Cohort. AIDS Res Hum Retroviruses 33(1): 41-48. (2017).
- Kourí V, Khouri R, Alemán Y, Abrahantes Y, Vercauteren J, Pineda-Piña AC, et al. CRF19_cpx is an evolutionary fit HIV-1 variant strongly associated with rapid progression to AIDS in Cuba. EbioMedicine 2: 244-254. (2015).