Information
Journal Policies
Opioid, Dopamine, and GABA Receptor Addiction and Mental Health Enriched Genomic Areas Co-Located with 16 Cancer, Immunity and Parkinson’s Disease Pharmacogenomics Drug Binding Sites
Latifa Jackson*
Copyright : © 2019. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
The molecular interactions between opioid, alcohol and dopamine class drug abuses and its co-morbidity with mental health conditions including schizophrenia, bipolar disorder and depression phenotypes have been the focus of intense interest. Genomic variants in genes contributing to addiction and mental health co-morbidities have been identified as contributing to the severity, duration and potential therapeutic outcomes for patient. Currently therapies for addiction and mental health arise from exhaustive molecular genomic analyses with subsequent investigational assessments. Our previous work demonstrates that using an in silico functional genomics and evolutionary biology framework can identify potential functional variants with import for opioid, dopamine and alcohol addiction complex polygenic traits. This approach may also be useful in identifying pharmacogenomics therapeutic targets at addiction and mental health genes. We sought to evaluate whether the intersection. NCBI curated genes identified as participating in addiction phenotypes along with schizophrenia, bipolar disorder and depression (N=1968 genes) were projected onto the human genome to find eight areas of addiction and mental health gene set enrichment. Functional annotations were conducted for core phenotype functions. Finally, drug binding annotations were conducted to determine whether the binding sites sitting in these addiction and mental health hotspots were related to the functional or gene set enrichment. We found that 16 pharmacogenomic drug binding sites were identified in this analysis which broadly fell into four categories: cancer, addiction, immunity and mental health phenotypes drugs. These findings suggest that addiction and immunity enriched gene areas are important to understanding therapeutic treatment options and that using this in silico approach can return results consistent with the gene set enriched hotspot annotation.
1. Introduction
Substance addiction disorders represent a major economic and health cost to human populations (Lynskey & Strang, 2013).Substance addiction is loosely defined as a chronic relapsing spectrum disorder characterized by loss of control over substance taking (Girard & Carlton, 1978; Goodman, 1990; Peele, 1977).Opiate, dopamine and GABA addictions are complex diseases with strong genetic components. These three substance disorders represent significant costs to the global judicial and healthcare systems. The treatment of addiction is further confounded by the co-occurrence of other pathologies that complicate treatment regimes (Crews, 2012; Uhl et al., 2008). For example, addiction and mental health are well-characterized co-morbidities. Mental health conditions such as depression, bipolar disorder and schizophrenia have clear genetic synergies between the prevalence of one mental health condition and addiction. This dissertation focuses on the characterization of addiction hotspots in the genome, their interplay with mental health genetics and then examines how infectious disease burden is correlated to the rise of immune and addiction variants. Of particular interest are the metabolic and addictive properties of opioid, GABA-receptor, and dopamine receptor class substances, that have been identified as exerting an oversize public health crisis effect on Americans(Stefano, Fricchione, Goumon, & Esch, 2005). Similarly, neurological disorders such as depression, bipolar disorder, and schizophrenia represent a significant strain on health and judicial entities. Both disorder classes have long been identified as co-morbidities (Regier et al., 1990). Both substance addiction and mental health are behavior-based phenomenon representing a diverse array of psychological, biological, and genetic attributes and environmental and cultural factors (Goodwin, 1975; Kamerow, Pincus, & Macdonald, 1986; Naranjo & Bremner, 1993; Tomkins & Sellers, 2001; Truan, 1993).
A number of studies have identified the clinical intersection of mental health and substance addiction traits (Blum, Oscar-Berman, Badgaiyan, Palomo, & Gold, 2014; McKetin et al., 2014). This has been shown in a diverse variety of mental health conditions such as depression (Jegede, 1978; Woody, O'Brien, & Rickels, 1975), bipolar disorder (Altamura, 2007; Geoffroy, Goddefroy, Rolland, & Cottencin, 2012; Maremmani, Perugi, Pacini, & Akiskal, 2006), and schizophrenia (Batel, 2000; Dubertret, Bidard, Ades, & Gorwood, 2006; Falkai & Moller, 2011; Laviolette & Grace, 2006; Schmidt & Beninger, 2006; Seibyl, Brenner, Krystal, Johnson, & Charney, 1992). To date, the crosstalk of addiction and mental health genetic contributors has not been fully understood based on association studies (Spanagel et al., 2010; Treutlein & Rietschel, 2011) and functional genomics (Ho et al., 2010; Vercauteren, Bergeron, Vandesande, Arckens,& Quirion, 2004). In this study we focus on the addiction disorders for opiate, dopamine, and GABA addiction as well as the mental health disorder identified as depression, bipolar disorder and schizophrenia. Of particular interest is the relationship between genes and variants with their pharmocogenomic drug targets.
2.Methods
A search of NCBI Gene based on strings of disease words was used in order to generate a list of genes with biological relevance to addiction. Genes for dopamine, opiate and GABA addiction; bipolar disorder; depression; and schizophrenia were identified. We mapped the resulting list of genes onto human chromosomes and considered the clusters they form. A Mat lab based program script was written to cluster genes in the genome and report those hotspot regions back as an output file. This Jackson hotspot approach enables the identification of candidate genes sitting adjacent to known addiction hotspot genes and the subsequent identification of the candidate polymorphisms in a diverse array of human populations(Jackson, 2017).
Hotspots were defined as genic regions approximately 2 Mb in length along the genome, which contained 13 or more genes identified from the combined addiction and mental health gene list. Additionally, we collapsed multiple small genomic hotpot windows into larger ones that spanned the 2Mb or smaller region. Each hotspot contained genes not currently associated with these joint addiction and mental health phenotypes. Genes were included in this analysis based on the high probability of common regulation patterns within a linear chromosomal region defined by the hotspot.
Genes located within hotspots were considered in two ways in statistical enrichments: all genes in the hotspot window, and only those previously linked to addiction. All genes in the hotspots were annotated using DAVIDs Bioinformatics Resources Tool software (Huang da, Sherman, & Lempicki, 2009; Huang da, Sherman, Zheng, et al., 2009) for biological process, molecular function, cell compartment and KEGG pathways (Kanehisa, 2002; Kanehisa, Goto, Kawashima, & Nakaya, 2002). Functional enrichments were quantified using Benjamini score analysis cutoffs of 0.01 (Benjamini, Drai, Elmer, Kafkafi, & Golani, 2001). Finally, all genes in hotspots were annotated for drug interactions using Drug Bank (Knox et al., 2011; Law et al., 2014; Wishart et al., 2008; Wishart et al., 2006).
Once hotspots were indentify.
3. Results
Literature curated addiction and mental health genes were mapped onto human chromosomes. Clusters of these genes identify hotspot locations within the genome. These hotspots contain genes previously not linked to addiction and mental health as well as regulatory motifs with drug binding sites. Our bioinformatics based discovery and subsequent investigation of combined addiction and mental health hotspots reveal their functional roles. Results point out multiple hotspots participating in dual addiction and mental health phenotypes. When addiction and mental health loci were jointly considered, hotspots were identified.
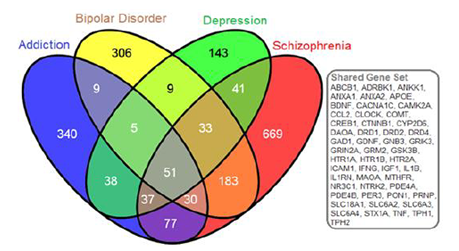
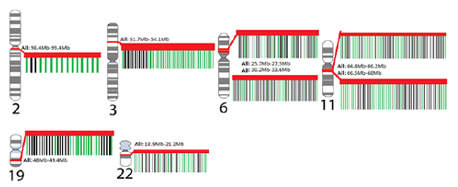
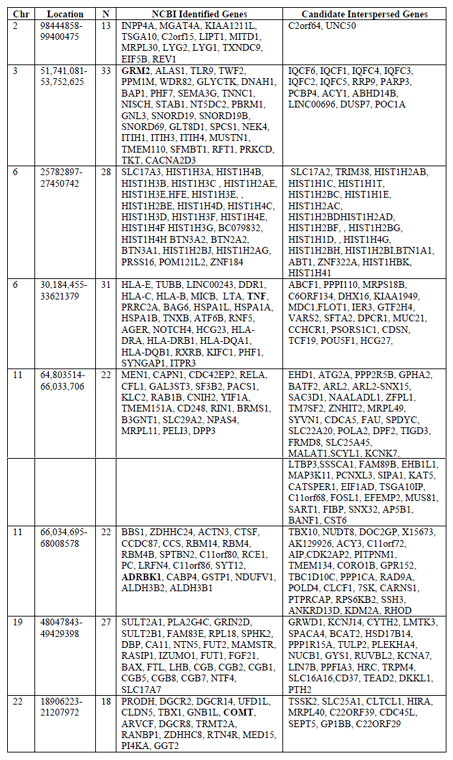
Gene searches were conducted to identify genes belonging to NCBI curated mental health and addiction genes lists. The bipolar disorder gene list comprised 626 genes, depression comprised 357 genes, schizophrenia comprised 1121 genes and Addiction comprised 587 genes garnered from opiate, GABA and dopamine addictions. When considered together this genes list represented 1968 genes encoding putative addiction and mental health targets. Figure 1 shows the overlap in genes from these four gene lists. Interestingly, there were 51 genes that were shared between all gene lists but only four of these shared genes were represented in the eight hotspots. These are indicated in red in Table 1. This overlap gene set contained the DRD, HTR and SLC6A gene families as well as a host of immune function genes including: ICAM1, IFNG, IGF1, IL1B, IL1RN, and TNF. Of this combined set of addiction and mental health genes, 192 genes fell into cluster regions within the genome. Our analyses identified these eight genomic regions with significant numbers of genes involved in the dual disorder. These eight regions are shown in Figure 2 with their associated NCBI-identified genes (green) and the candidate genes (black) that are interspersed. Hotspots ranged from having 13-33 literature curated genes in a genomic window approximately 2Mb or smaller.
When genes were mapped to the genome, we found eight regions smaller than 1.5 Mb each with 11- 38 genes identified from NCBI Gene curated gene lists. Additionally we consider the genes interspersed with our set of addiction and mental health genes. Red gene names are those that were among the 51 genes shared by all addiction and mental health disorders.
The Venn diagram provides the gene symbols (N=1968 genes) for intersections of gene sets corresponding to dopamine, opiate, and GABA addictions with bipolar disorder, depression, and schizophrenia. The gray boxed genes are those genes (n=51) common to all disorders.
Each karyotype of a chromosome shows the location of the genes that are represents as hotspots. Genes that lie within each hotspot were identified through USCS Genome Browser. The insets for each hotspot show genes identifies curated genes (green) and interspersed genes (black) with a red band showing the entire hotspot region.
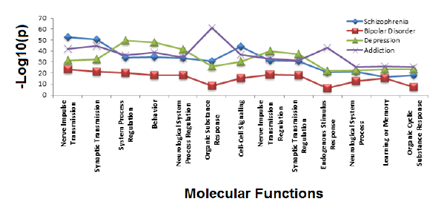
A functional annotation on the complete list of NCBI identified genes was performed to determine the major functional roles for addiction and mental health genes situated in the genomic enrichment region. Functional annotation of the combined set of depression, schizophrenia, bipolar and addiction genes found commonalities as identified in Figure 3, with shared function revolving around neurological function, response organic substances and cell-cell signaling. Addiction and depression overlapped in three functional regulatory roles: cellular localization, cyclic nucleotide biosynthesis and metabolism. Additionally when pair wise comparisons were made, depression and schizophrenia gene lists overlapped in homeostatic processes, while bipolar and schizophrenia genes jointly participated in neuronal development and cellular differentiation. Hotspot gene lists were used to identify gene ontology biological processes and molecular functions enriched for each hotspot. When we considered the identified literature curated genes that identified each hotspot and then those genes that were not previously identified, but could be addiction and mental health candidates, we found no real differences in the functional annotation of these sets.
The top 25 significant functional annotations were obtained (Benjamini > 0.01) for gene ontology molecular function for each addiction, bipolar, depression, schizophrenia gene class. The vertical axis of each graph shows the –log10 (p) while the horizontal axis indicates the functional annotations. The gene lists are color coded for depression (green), bipolar disorder (red), schizophrenia (blue). Addiction genes are a composite of opiate, GABA, and dopamine addiction genes (purple). Thirteen functions are shared between the gene sets.
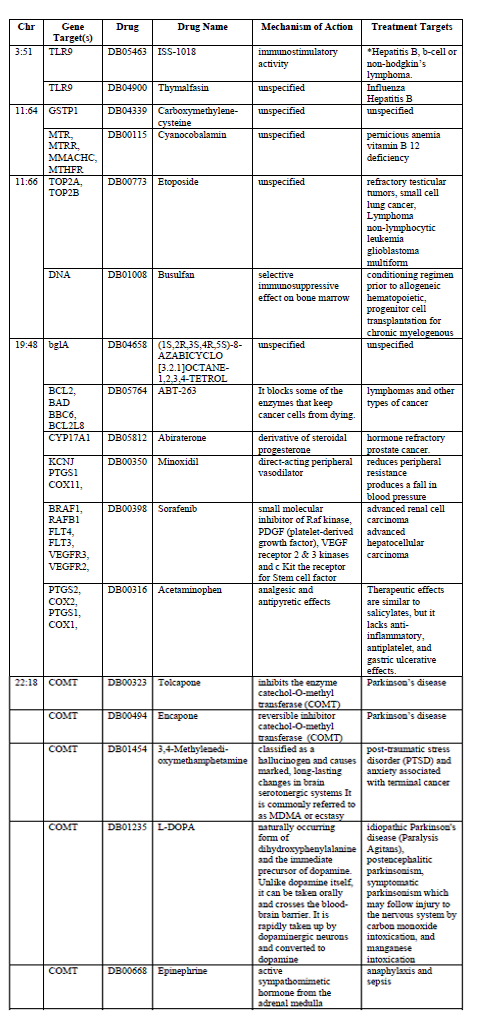
Drug target annotation was performed to determine the role that pharmacological products interacted to binding sites within these hotspots. When the hotspot regions were annotated for drug interactions 16 drugs were found to have binding sites. Addiction, bipolar, depression and schizophrenia drug interactions are characterized in Table 2.
While not all hotspots had drug binding sites located within their domains, five of the hotspots did. When identified by the types of drugs found at these five hotspots we found some thematic divisions in drug targets. Five drug binding sites are associated with cancer therapies: Sorafenib, Abiraterone, ABT-263, Etoposide, and Thymalfasin. Four drugs were associated with the mental health condition Parkinson’s disease, post-traumatic stress disorder or anxiety: L-DOPA, 3,4-Methylenedi-oxymethamphetamine, Encapone, and Tolcapone. One drug binding sites was specifically targeted ecstasy.
4. Discussion
We have observed that addiction and mental health genes create eight hotspots in the genome. Their shared functional annotation and drug binding site annotation support the idea that this clustering is meaningful. This approach helps to unify the disparate clinical, genetic, and functional observations about addiction and mental health co-morbidity. Our analyses demonstrate that genes curated for their involvement in opiate, GABA and dopamine addictions share significant genomic position overlap with genes involved in the bipolar disorder, depression and schizophrenia. Additionally when we perform functional annotation on these gene sets, we find that they share core molecular processes such as cell-cell signaling, synaptic transmission and responses to organic substances. These genes also share more amorphous processes such as learning, memory and behavior. Finally we found that when these addiction and mental health genetic hotspots were annotated for their drug interactions, they identified binding sites for illicit drugs, cancer drugs, and neurological degenerative disorders such as Parkinson’s disease.
While there has been significant identification of the clinical synergies between addiction and some mental health phenotypes (Kamerow et al., 1986; Tomkins & Sellers, 2001; Uhl et al., 2008), we show here that a subset of these addiction and mental health genes actually sit together in genomic windows. These genomic windows contain four of the 51 genes that are common to all gene lists (Table 1); instead we found that genes sitting in genomic hotspots were often shared by only two of the comparisons. When mapped to the genome, these regions were not random with respect to genome locality. These eight hotspot regions contain 192 of the 1968 unique genes identified as participating in addiction and mental health genetics and represent nearly 10% of genes associated with addiction and mental health. A number of studies have identified the clinical intersection of mental health and substance addiction traits (Blum et al., 2014; McKetin et al., 2014). This has been shown in a diverse variety of mental health conditions such as depression (Jegede, 1978; Woody et al., 1975), bipolar disorder (Altamura, 2007; Geoffroy et al., 2012; Maremmani et al., 2006), and schizophrenia (Batel, 2000; Dubertret et al., 2006; Falkai & Moller, 2011; Laviolette & Grace, 2006; Schmidt & Beninger, 2006; Seibyl et al., 1992). To date, the crosstalk of addiction and mental health genetic contributors has not been fully understood based on association studies (Spanagel et al., 2010; Treutlein & Rietschel, 2011) and functional genomics (Ho et al., 2010; Vercauteren et al., 2004).
Functional annotation of genes residing in the windows provides a verification of the role that these genes play within the wider context of the genomic region they inhabit. It is reassuring that our analyses return functional categories emphasizing the shared role that neurological system regulation, cell-cell signaling, stimulus response and organic substance response play in the development of mental health and addiction disorders. These findings support the complex functional interactions between opiate, dopamine and GABA addictions with schizophrenic, depressive and bipolar mental health disorders.
Previous studies have identified that drugs involved with addiction and mental health may have pleiotropic effects (Uhl et al., 2008). It is therefore of significant interest to characterize the drug targets in these hotspot regions. We found that drug binding sites fall in to three major therapeutic categories, those related to cancers, those related to up regulation of immune responses and those involved in neurodegenerative disorders. Of particular interest was the hotspots located on chromosome 19 (ABT-263, Abiraterone, Minoxidil, and Sorafenib) and chromosome 22 (Acetaminophen, Tolcapone, Encapone, 3,4-Methylenedi-oxymethamphetamine, L-DOPA and Epinephrine).
The observation of increased anxiety behaviors and morphine consumption in a Sprague-Dawley rat model suggests that genetic variation in the epinephrine-mediated norepinephrine signaling pathway may be a novel mechanism for affective behavior such as anxiety and addiction(Brody et al., 2014). In a human study of African American young adults, levels of urinary epinephrine were predictive of drug use a year later( Zhai& Sun, 2013). The drug binding site for epinephrine is located in the chromosome 22 hotspot gene COMT. Sorafenib, a hepatocellular carcinoma treatment, is mediated by cellular signaling mechanisms. This drug target annotation is consistent with the genomic functional annotation of the genes underlying addiction and mental health disorders. There is some evidence of genetic variant mediated drug resistance. Sorafenib can activate addiction switches leading to reduced drug efficacy (McElligott et al., 2013). While the mechanisms of this are not fully elucidated, the localization of Sorafenib among known addiction genes could be a reason for this trigger and bears further investigation.
This Jackson Hotspot Approach could identify genomic sites that sat at the intersection of addiction to opiate, GABA and dopamine with mental health disorders identified as depression, schizophrenia, and bipolar disorder. We have shown that genes involved separately in these two disorders are co-located at eight genomic regions. Additionally we hypothesized that each of these regions might have drug binding sites that share functional annotations with the genes identified in the region. Our analyses have identified 16 drug binding target regions located in five of the eight hotspot regions which share functional or therapeutic activity with addiction and mental health disorder phenotypes. This finding compels us to speculate on the role that functional ontology plays in the primary of counter-indicative phenotypes that these drugs present.
References
- Altamura, A. C. (2007). Bipolar spectrum and drug addiction. J Affect Disord, 99(1-3), 285. doi:10.1016/j.jad.2006.09.005
- Batel, P. (2000). Addiction and schizophrenia. Eur Psychiatry, 15(2), 115-122.
- Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N., & Golani, I. (2001). Controlling the false discovery rate in behavior genetics research. Behav Brain Res, 125(1-2), 279-284.
- Blum, K., Oscar-Berman, M., Badgaiyan, R. D., Palomo, T., & Gold, M. S. (2014). Hypothesizing dopaminergic genetic antecedents in schizophrenia and substance seeking behavior. Med Hypotheses, 82(5), 606-614. doi:10.1016/j.mehy.2014.02.019
- Brody, G. H., Yu, T., Mackillop, J., Miller, G. E., Chen, E., Obasi, E. M., & Beach, S. R. (2014). Catecholamine levels and delay discounting forecast drug use among African American youths. Addiction. doi:10.1111 /add.12516
- Crews, F. T. (2012). Immune function genes, genetics, and the neurobiology of addiction. Alcohol Res, 34(3), 355-361.
- Dubertret, C., Bidard, I., Ades, J., & Gorwood, P. (2006). Lifetime positive symptoms in patients with schizophrenia and cannabis abuse are partially explained by co-morbid addiction. Schizophr Res, 86(1-3), 284-290. doi:10.1016/ j.schres.2006.05.006
- Falkai, P., & Moller, H. J. (2011). The functional sequelae of schizophrenia: consequences of long-term pharmacotherapy and the neurobiology of addiction. Eur Arch Psychiatry Clin Neurosci, 261(2), 83-84. doi:10.1007/s00406-011-0190-x
- Geoffroy, P. A., Goddefroy, G., Rolland, B., & Cottencin, O. (2012). Efficacy of aripiprazole in comorbid addiction in bipolar disorder. CNS Neurosci Ther, 18(4), 359-360. doi:10.1111 /j.1755-5949.2012.00301.x
- Girard, D. E., & Carlton, B. E. (1978). Alcoholism. Earlier diagnosis and definition of the problem. West J Med, 129(1), 1-7.
- Goodman, A. (1990). Addiction: definition and implications. Br J Addict, 85(11), 1403-1408.
- Goodwin, D. W. (1975). Genetic determinants of alcohol addiction. Adv Exp Med Biol, 56, 339-355.
- Ho, M. K., Goldman, D., Heinz, A., Kaprio, J., Kreek, M. J., Li, M. D., . . . Tyndale, R. F. (2010). Breaking barriers in the genomics and pharmacogenetics of drug addiction. Clin Pharmacol Ther, 88(6), 779-791. doi:10. 1038/clpt.2010.175
- Huang da, W., Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4(1), 44-57. doi:10.1038/ nprot.2008.211
- Huang da, W., Sherman, B. T., Zheng, X., Yang, J., Imamichi, T., Stephens, R., & Lempicki, R. A. (2009). Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics, Chapter 13, Unit 13 11. doi:10.1002/0471250953.bi1311s27
- Jackson, L. F. (2017). A Novel Bioinformatic Approach to Understanding Addiction. Journal of Family Strengths, 7(1), 4.
- Jegede, R. O. (1978). Depressive symptomatology in drug addicts: therapeutic and aetiological implications. Afr J Med Med Sci, 7(3), 171-174.
- Kamerow, D. B., Pincus, H. A., & Macdonald, D. I. (1986). Alcohol abuse, other drug abuse, and mental disorders in medical practice. Prevalence, costs, recognition, and treatment. JAMA, 255(15), 2054-2057.
- Kanehisa, M. (2002). The KEGG database. Novartis Found Symp, 247, 91-101; discussion 101-103, 119-128, 244-152.
- Kanehisa, M., Goto, S., Kawashima, S., & Nakaya, A. (2002). The KEGG databases at GenomeNet. Nucleic Acids Res, 30(1), 42-46.
- Knox, C., Law, V., Jewison, T., Liu, P., Ly, S., Frolkis, A., . . . Wishart, D. S. (2011). DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res, 39(Database issue), D1035-1041. doi:10.1093/ nar/gkq1126
- Laviolette, S. R., & Grace, A. A. (2006). The roles of cannabinoid and dopamine receptor systems in neural emotional learning circuits: implications for schizophrenia and addiction. Cell Mol Life Sci, 63(14), 1597-1613. doi:10.1007/s00018-006-6027-5
- Law, V., Knox, C., Djoumbou, Y., Jewison, T., Guo, A. C., Liu, Y., . . . Wishart, D. S. (2014). DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res, 42(Database issue), D1091-1097. doi:10.1093/nar/gkt1068
- Lynskey, M. T., & Strang, J. (2013). The global burden of drug use and mental disorders. Lancet, 382(9904), 1540-1542. doi:10.1016/ S0140-6736(13)61781-X
- Maremmani, I., Perugi, G., Pacini, M., & Akiskal, H. S. (2006). Toward a unitary perspective on the bipolar spectrum and substance abuse: opiate addiction as a paradigm. J Affect Disord, 93(1-3), 1-12. doi:10.1016/j.jad.2006.02.022
- McElligott, Z. A., Fox, M. E., Walsh, P. L., Urban, D. J., Ferrel, M. S., Roth, B. L., & Wightman, R. M. (2013). Noradrenergic synaptic function in the bed nucleus of the stria terminalis varies in animal models of anxiety and addiction. Neuropsychopharmacology, 38(9), 1665-1673. doi:10.1038/npp.2013.63
- McKetin, R., Lubman, D. I., Najman, J. M., Dawe, S., Butterworth, P., & Baker, A. L. (2014). Does methamphetamine use increase violent behaviour? Evidence from a prospective longitudinal study. Addiction, 109(5), 798-806. doi:10.1111/add.12474
- Naranjo, C. A., & Bremner, K. E. (1993). Behavioural correlates of alcohol intoxication. Addiction, 88(1), 25-35.
- Peele, S. (1977). Redefining addiction. I. Making addiction a scientifically and socially useful concept. Int J Health Serv, 7(1), 103-124.
- Regier, D. A., Farmer, M. E., Rae, D. S., Locke, B. Z., Keith, S. J., Judd, L. L., & Goodwin, F. K. (1990). Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. JAMA, 264(19), 2511-2518.
- Schmidt, W. J., & Beninger, R. J. (2006). Behavioural sensitization in addiction, schizophrenia, Parkinson's disease and dyskinesia. Neurotox Res, 10(2), 161-166.
- Seibyl, J. P., Brenner, L., Krystal, J. H., Johns on, R., & Charney, D. S. (1992). Mazindol and cocaine addiction in schizophrenia. Biol Psychiatry, 31(11), 1179-1181.
- Spanagel, R., Bartsch, D., Brors, B., Dahmen, N., Deussing, J., Eils, R., . . . Zimmer, A. (2010). An integrated genome research network for studying the genetics of alcohol addiction. Addict Biol, 15(4), 369-379. doi:10.1111/ j.136 9-1600.2010.00276.x
- Stefano, G. B., Fricchione, G., Goumon, Y., & Esch, T. (2005). Pain, immunity, opiate and opioid compounds and health. Med Sci Monit, 11(5), MS47-53.
- Tomkins, D. M., & Sellers, E. M. (2001). Addiction and the brain: the role of neurotran smitters in the cause and treatment of drug dependence. CMAJ, 164(6), 817-821.
- Treutlein, J., & Rietschel, M. (2011). Genome-wide association studies of alcohol dependence and substance use disorders. Curr Psychiatry Rep, 13(2), 147-155. doi:10.1007/s11920-011-0176-4
- Truan, F. (1993). Addiction as a social construction: a postempirical view. J Psychol, 127(5), 489-499. doi:10.1080/00223980.1993. 9914886
- Uhl, G. R., Drgon, T., Johnson, C., Li, C. Y., Contoreggi, C., Hess, J., . . . Liu, Q. R. (2008). Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify "connectivity constellation" and drug target genes with pleiotropic effects. nn N Y Acad Sci, 1141, 318-381.doi:10.119 6/annals.1441.018
- Vercauteren, F. G., Bergeron, J. J., Vandesande, F., Arckens, L., & Quirion, R. (2004). Proteomic approaches in brain research and neuropharmacology. Eur J Pharmacol, 500(1-3), 385-398. doi:10.1016/j.ejphar.2004. 07.039
- Wishart, D. S., Knox, C., Guo, A. C., Cheng, D., Shrivastava, S., Tzur, D., . . . Hassanali, M. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res, 36(Database issue), D901-906. doi:10.10 93/nar/gkm958
- Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., . . .Woolsey, J. (2006). DrugBank: a comprehen sive resource for in silico drug discovery and exploration. Nucleic Acids Res, 34(Database issue), D668-672. doi:10.1093/ nar/gkj067
- Woody, G. E., O'Brien, C. P., & Rickels, K. (1975). Depression and anxiety in heroin addicts: a placebo-controlled study of doxepin in combination with methadone. Am J Psychiatry, 132(4), 447-450.
- Zhai, B., & Sun, X. Y. (2013). Mechanisms of resistance to sorafenib and the corresponding strategies in hepatocellular carcinoma. World J Hepatol, 5(7), 345-352.doi:10.4254/wjh.v5.i7. 345