Information
Journal Policies
Opioid Metabolizing Enzyme Allele Frequencies and Drug Use in a Cohort of African American Young Adults
Bradford D.Wilson1,Earl B.Ettienne2,Victor Apprey3,Adaku Ofeogbu4,Muneer Abbas5,Georgia M.Dunston6,Forough Saadatmand7
2.College of Pharmacy, Howard University 2300 4th St., NW Washington DC 20059.
3.Department of Microbiology, Howard University College of Medicine, 520 W St. Washington DC 20059.
4.Department Pediatrics and Child Health, Howard University College of Medicine 2041 Georgia Ave, NW Washington DC, 20060
Copyright : © 2017 . This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Background:The current opioid epidemic has become a top priority of our national research institutions. Medication assisted therapy (MAT) is an effective treatment for opioid use disorder (OUD). The use of pharmacogenetic testing to guide dosing of medications for OUD has been reported to improve MAT outcomes. The population used to develop pharmacogenetic panels lacked diversity. We determined functional variants in opioid metabolizing enzymes and tested for association with drug use in a cohort of African American young adults.
Participants and Methods: We genotyped four SNPs in CYP2B6, CYP2D6, and CYP3A4 in a cohort of 575 African American young adults and tested for association with drug use by logistic and linear regression.
Results: TheCYP2D6*17 allele (rs28371706) is associated with other drug use(OR = 7.90, 95% CI:1.05-59.22, p=0.05968)and the CYP2D6*29 (rs59421388)allele is associated with other drug use over the past thirty days under the dominant(p = 0.0008681), co dominant (p = 0.0046255), and over-dominant (p = 0.0042272) models.
Conclusion: In this cohort, use other of drugs like heroin, LSD, PCP, and MDMA may be associated with the CYP2D6*17 and CYP2D6*29 alleles. Additional study is needed to further explore these findings.
Pharmacogenetics, opioids, addiction, African Americans,Addiction
1. Introduction
Opioid use disorder (OUD) carries a significant public health burden, with 12.5 million people reporting misuse of prescription pain relievers and over 300,000 people reporting heroin use in 2015 [1].The 200%increase in over dose deaths since 2000 has elevated opioid addiction to the epidemic level, as the leading cause of unintentional injury death in the US[2],[3]. The recent drastic increase in overdose deaths has been attributed, in large part, to the relaxation of regulations on prescribing opioids for chronic non-cancer pain [2],[4].The rise in the number of opioid prescriptions and subsequent long-term use of opioids has significantly contributed to the problem with regards to increased access and use. Addiction is complex and Is defined a primary, chronic disease of brain reward, motivation, memory and related circuitry, dysfunction in which leads to characteristic biological, psychological, social and spiritual manifestations [5].The reward pathway has been of interest with regards to the role of dopamine and dopamine receptors [6],[7]. Pharmaco - genomic (PGx) variation has also been hypothesized to play a role in the biology of addiction via the facilitation of tolerance [8].
Variants have been identified in all of the members of the CYP450 enzyme family genes [9]-[14]. However, the populations in which these variants were identified lacked diversity. African diaspora populations are known to have higher levels of genetic diversity than non-African populations [15],[16]. The under representation of populati-ons of African descent in early research on the CYP450 gene family has resulted in a deficiency of research on the full spectrum of functionally significant variation in drug metabolizing enzyme genes. There is a need for more comprehensive functional characterization of CYP450 gene variation that can only arise from increased study population diversity. We recently reported a case study on the use of PGx-guided buprenorphine dosing for medication assisted therapy (MAT) in OUD involving CYP3A4*1B allele genotyping [17]. According to 1000 Genomes Project data, the variant allele (G) has a wide frequency distribution ranging from 0% in Chinese populations to up to 84% in West African populations.CYP3A4*1B has been reported to dominantly confer an ultra rapid metabolize (UM)phenotype [17],[18].In this study, we characterize allelic frequencies of known functional variants in opioid metabolizing CYP450 genes in a population of African American young adults and explore the relationship between these variants and drug use in the cohort.
2. Materials & Methods
This study stems from a parent project entitled ―Biological and Social Correlates of Drug Use in African American Emerging Adults‖ (BADU). The BADU dataset is composed of 557 native-born African-American young adults aged 18-25recruited in Washington D.C from 2010 to 2012 and contains information on 274 females and 283 males.
Data on family history of alcohol and drug use and study participant alcohol, tobacco, and other drug (ATOD) use were collected via the Family Tree Questionnaire (FTQ) and Centers for Disease Control (CDC) Youth Risk Behavior Survey (YRBS) instruments respectively. The drug use questions from these instruments are listed in appendix 1.Data from responses to all questions on drug and opioid use in these two surveys was analyzed.
Deoxyribonucleic acid (DNA) collected from participants was genotyped to test opioid drug metabolizing enzyme (DME) variants for association with drug use. Genotyping was performed using the Applied Bio systems 7900 HT Fast Real-Time PCR System. The dried-down DNA method was employed for sample preparation. Briefly, to each dried DNA sample, 2.5uL of 2X TaqMan® master mix, 0.25 uL of 20X assay uL working stock, and 2.25 uL of nuclease-free water were added for a total reaction volume of 5.0 uL according to the manufacturer’s protocol. The genotyped SNPs listed in Table 1were selected based on reported functional significance in populations of African descent. They were genotyped using TaqMan® Drug Metabolism Genotyping Assay numbers; C_60732328_20, C_2222771_40, C_348161 13_20, C_27859822_10, and C_30634202_10 which genotype SNPsrs 28399 499, rs28371706, rs59421388, rs4987161, and rs12721629 respectively.
Statistical Analysis: The association of other drug use and other drug use over the past 30 days with SNP genotype was analyzed by logistic and linear regression respectively using the SPSS and R statistical software programs [19],[20]. For this study, the major allele found in people of African descent was considered the reference allele. For each SNP genotype the dominant, co-dominant, recessive and log-additive genetic models were tested. Confidence intervals (CIs) of 95% were set on the calculated odds ratios (ORs). Two sided P-values of < 0.05 were considered to be statistically significant. The adjustment for multiple comparisons was made using the Bonferroni test.
3. Results & Discussion
The observed frequencies of the opioid DME SNPs are also listed in Table 1.
The frequencies of the four alleles in our cohort was comparable to the frequencies reported by the SNP genotyping assay manufacturer. SNP rs28399499 is reported at frequencies of 0.08% and 0.12% in the 1000 Genome African (AFR) and HapMap Yoruba (YRI) populations respectively. We found an association between the CYP2D6*17allele (rs28371706)and other drug (heroin, LSD, PCP, Ecstasy, etc.) use (OR = 7.90, 95% CI: 1.05-59.22, p=0.05968) under the recessive model and between the CYP2D6*29allele (rs59421388) and other drug use over the past 30 days under the dominant (β = 0.8462,p = 0.0008681, 95% CI: -1.262 - - 0.4300),co dominant (β = 0.8462,p =0.00462 55,95% CI: -1.297 - -0.3953), and over dominant (Β = 0.7857, p = 0.0042272,95% CI: - 1.256 - -0.3152) models as shown in Tables 2 & 3 respectively.
The dominant model is optimal with the lowest Akaike information criterion (AIC) value of 29.00.The log additive model p value of 0.0023699 indicates that there is a statistically significant difference between the genotypes. The linear regression also revealed an associ-ation between CYP2D6*29(Pr(>|t|) = 0.0234) and other drug use over the past 30 days after adjusting for the other four SNPs as shown in Table 4
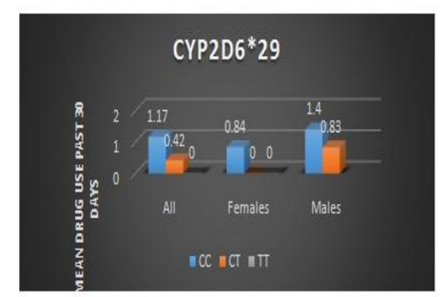
This association was even stronger when we analyzed females only (Pr(>|t|) = 0.00748) and is shown in Table 5.Figure 1 illustrates the increased average drug use associated with the C allele.
The CYP2D6*17(rs28371706) and CYP2D6*29 (rs59421388) alleles both code for mis sense transition mutations that result in decreased enzymatic activity [21],[22]. These alleles have also been shown to exhibit changes in substrate specificity, in metabolizingdebrisoquine and dextromethorphan significantly slower than codeine or metoprolol [23]. Slow or reduced drug metabolism has been hypothesized to predispose to tolerance, a posited precursor of dependence and addiction [8]. Although there are reports on the role of impaired drug metabolism in the development of addiction, there are limitations to this study.
The sample size is small with regards to the number of individuals who had tried other drugs (heroin, LSD, PCP, Ecstasy, etc.) (N= 61). The survey items in the instruments used to collect data on drug use did not differentiate opiates from other classes of drugs including LSD, MDMA, and PCP in assessing history of family and participant history of other drug use. For participants who responded affirmatively to the questions on other drug use who were referring to opioids, there was no data collected on the specific opiate of choice (e.g. codeine, dextro methorphan, fentanyl, heroine, or oxycodone). Given the complexity of the addiction phenotype, it is likely that there are a number of genetic factors contributing to the biology underlying opioid addiction. Variants in drug metabolizing enzymes, dopamine and opioid receptors, and reward pathway anomalies may all play a role.
4. Conclusion
We found an association between the CYP2D6 *17 and *29 alleles and other drug use in our population. A larger study that specifi- cally identifies the drugs being used and obtains additional genetic data is needed to further explore these finding and validate the asso-ciation. Additional study of these variants in an OUD population are needed to explore the relationship of these alleles to addiction.
5.Acknowledgement
―Research reported in this publication was supported by the National Institute on Drug Abuse of the National Institutes of Health under Award Number R24DA021470. The data used in this study was collected under Dr. Kathy Sanders-Phillips that was supported by National Institute of Minority Health and Health Dispar-ity (grant #P20MD000198) and the NIH ―Re-Engineering the Clinical Research Enterprise,‖ (Grant # UL1TR000101). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.‖
"This project was also supported in part by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number G12MD007597." to be inserted immediately before "The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health."
References
- U.S. Department of Health and Human Services (HHS), Office of the Surgeon General, Facing Addiction in America:The Surgeon General’s Report on Alcohol, Drugs, and Health. Washington, DC: HHS, November 2016.
- Rudd R., Aleshire N., Zibbell J., Gladden R., Increases in Drug and Opioid Overdose Deaths— United States, 2000–2014, American Journal of Transplantation. 16, 1323–1327 (2016)
- National Academies of Sciences, Engineering, and Medicine. 2017. Pain Management and the Opioid Epidemic: Balancing Societal and Individual Benefits and Risks of Prescription Opioid Use. Washington, DC: The National Academies Press. https://doi.org/10.17226/24781.
- 4.Manchikanti L., Helm II S., Fellows B., Janata J., Pampati V., Grider J.,Boswell M., Opioid Epidemic in the United States, Pain Physician; 15 ES9-ES38 (2012)
- Smith, DE. Editor's Note: The Process Addictions and the New ASAM Definition of AddictionJournal of Psychoactive Drugs Vol. 44, Iss. 1,2012
- Comings DE1, Blum K. Reward deficiency syndrome: genetic aspects of behavioral disorders. Prog Brain Res. 2000; 126: 325-41.
- Foll, Bernard Le; Gallo, Alexandra; Strat, Yann Le; Lu, Lin; Gorwood, Philip. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behavioural Pharma cology: February 2009 - Volume 20 - Issue 1 - pp 1-17doi: 10.1097/ FBP. 0b013 e3283242f05
- Kreek MJ1, Bart G, Lilly C, LaForge KS, Nielsen DA: Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev.2005 Mar;57(1):1-26.
- Pinto N, and Dolan M: Clinically Relevant Genetic Variations in Drug Metabolizing Enzymes. Curr Drug Metab 2011, 12(5): 487-49
- Xie HG, Kim RB, Wood AJ, Stein CM:Molecular basis of ethnic differences in drug disposition and response. Annu Rev PharmacolToxicol200141:815-850
- Singh D, Kashyap A, Pandey RV, Saini KS:Novel advances in cytochrome P450 research. Drug Discov Today 2011 16(17-18):793-9.
- Ingleman- Sundberg M: Polymorphism of Cytochrome P450 and Xenobiotic Activity. Toxicology 2002181-182:447-452
- Ingleman-Sundberg M: Genetic Susceptibility to Adverse Efects of Drugs and Environmental toxicants; The Role of the CYP Family. Mutation Research 2001482:11-19
- Scott S: Personalizing Medicine with Clinical Pharmacogenetics. Genet Med 201112:987-995
- Tishkoff SA, Williams SM: Genetic analysis of african populations: human evolution and complex disease. Nat Rev 2002, 3:611-621
- Tishkoff SA, Verrelli BC:Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet20034:293-340
- Pharmacogenomics-guided policy in opioid use disorder (OUD) management: An ethnically-diverse case-based approach. Earl B. Ettienne, Edwin Chapman, Mary Maneno, Adaku Ofoegbu, Bradford Wilson, Beverlyn Settles-Reaves, Melissa Clarke, Georgia Dunston, Kevin Rosenblatt. Addictive Behaviors Reports 2017 Dec; 6: 8–14
- Westlind- Johnsson, A., et al.: Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin Pharmacol Ther, 2006. 79(4): p. 339-49.
- IBM Corp. Released 2016. IBM SPSS Statistics for Windows, Version 24.0. Armonk, NY: IBM Corp.
- R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
- Oscarson M, Hidestrand M, Johansson I, Ingelman-Sundberg M: A combination of mutations in the CYP2D6*17 (CYP2D6Z) allele causes alterations in enzyme function. Mol Pharmacol. 1997 Dec; 52(6):1034-40.
- Wennerholm A, Johansson I, Hidestrand M, Bertilsson L, Gustafsson LL, Ingelman-Sundberg M: Characterization of the CYP2D6*29 allele commonly present in a black Tanzanian population causing reduced catalytic activity. Pharmacogenetics. 2001 Jul;11(5):417-27.
- Wennerholm A1, Dandara C, Sayi J, Svensson JO, Abdi YA, Ingelman-Sundberg M,Bertilsson L, Hasler J, Gustafsson LL: The African-specific CYP2D617 allele encodes an enzyme with changed substrate specificity. Clin Pharmacol Ther. 2002 Jan;71(1):77-88